Heart Disease Demo: How much risk for a single patient has
The previous post explained, overall, what factors affect health of the heart. This is the global interpretability of machine learning. A better tool for a subject matter expert to try out is on the boundary conditions. Here, he/she can put values which even confuses the experts; and see how well the model behaves. This is called local interpretability. This post is an example of local interpretability and how the model behaves well and erratically, given the input conditions.
The method used here is the use of SHAPELY values. To get an idea how this works, think of a game where each team member contributes to the final score.
A note on the parameters used in the demo
- Age: Age completed in years
- Resting blood pressure : Level of blood pressure at resting mode in mm/HG (Systoloc)
- Cholesterol: Serum cholesterol in mg/dl
- Maximum Heart Rate Achieved: Heart rate achieved while doing a treadmill test or exercise
- ST_Depression/oldpeak: Exercise induced ST-depression in comparison with the state of rest
- Sex: Gender of patient (The data had only male and female)
- Chest Pain Type: Type of chest pain experienced by patient
- Fasting blood sugar: Blood sugar levels on fasting > 120 mg/dl represents as 1 in case of true and 0 as false
- Resting ecg: Result of electrocardiogram while at rest
- Exercise angina: Angina induced by exercise 0 depicting NO 1 depicting Yes
- ST slope: ST segment measured in terms of slope during peak exercise
Try it out
(If you are loading this for first time, click on show widgets below, to load the application. Best viewed in bigger screen)
#HIDDEN
import pandas
import shap
import joblib
import ipywidgets as widgets
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.patches as mpatches
from sklearn.base import TransformerMixin, BaseEstimator
from pandas import Categorical, get_dummies
import time
from ipywidgets import interact, interact_manual,interactive
from ipywidgets import Layout, Button, Box, VBox
from ipywidgets import Button, HBox, VBox
from ipywidgets import Layout, Button, Box, FloatText, Textarea, Dropdown, Label, IntSlider
import warnings
warnings.filterwarnings("ignore")
#HIDDEN
class CategoricalPreprocessing(BaseEstimator, TransformerMixin):
def __get_categorical_variables(self,data_frame,threshold=0.70, top_n_values=10):
likely_categorical = []
for column in data_frame.columns:
if 1. * data_frame[column].value_counts(normalize=True).head(top_n_values).sum() > threshold:
likely_categorical.append(column)
try:
likely_categorical.remove('st_depression')
except:
pass
return likely_categorical
def fit(self, X, y=None):
self.attribute_names = self.__get_categorical_variables(X)
cats = {}
for column in self.attribute_names:
cats[column] = X[column].unique().tolist()
self.categoricals = cats
return self
def transform(self, X, y=None):
df = X.copy()
for column in self.attribute_names:
df[column] = Categorical(df[column], categories=self.categoricals[column])
new_df = get_dummies(df, drop_first=False)
# in case we need them later
return new_df
#HIDDEN
feature_model=joblib.load('random_forest_heart_model_v2')
categorical_transform=joblib.load('categorical_transform_v2')
explainer_random_forest=joblib.load('shap_random_forest_explainer_v2')
numerical_options=joblib.load('numerical_options_dictionary_v2')
categorical_options=joblib.load('categorical_options_dictionary_v2')
ui_elements=[]
style = {'description_width': 'initial'}
for i in numerical_options.keys():
minimum=numerical_options[i]['minimum']
maximum=numerical_options[i]['maximum']
default=numerical_options[i]['default']
if i!='st_depression':
ui_elements.append(widgets.IntSlider(
value=default,
min=minimum,
max=maximum,
step=1,
description=i,style=style)
)
else:
ui_elements.append(widgets.FloatSlider(
value=default,
min=minimum,
max=maximum,
step=.5,
description=i,style=style)
)
for i in categorical_options.keys():
ui_elements.append(widgets.Dropdown(
options=categorical_options[i]['options'],
value=categorical_options[i]['default'],
description=i,style=style
))
interact_calc=interact.options(manual=True, manual_name="Calculate Risk")
#HIDDEN
def get_risk_string(prediction_probability):
y_val = prediction_probability* 100
text_val = str(np.round(y_val, 1)) + "% | "
# assign a risk group
if y_val / 100 <= 0.275685:
risk_grp = ' low risk '
elif y_val / 100 <= 0.795583:
risk_grp = ' medium risk '
else:
risk_grp = ' high risk '
return text_val+ risk_grp
def get_current_prediction():
current_values=dict()
for element in ui_elements:
current_values[element.description]=element.value
feature_row=categorical_transform.transform(pandas.DataFrame.from_dict([current_values]))
feature_row=feature_row[['age', 'resting_blood_pressure', 'cholesterol',
'max_heart_rate_achieved', 'st_depression', 'sex_female', 'sex_male',
'chest_pain_type_non-anginal pain', 'chest_pain_type_asymptomatic',
'chest_pain_type_atypical angina', 'chest_pain_type_typical angina',
'fasting_blood_sugar_0', 'fasting_blood_sugar_1', 'rest_ecg_normal',
'rest_ecg_ST-T wave abnormality',
'rest_ecg_left ventricular hypertrophy', 'exercise_induced_angina_0',
'exercise_induced_angina_1', 'st_slope_flat', 'st_slope_upsloping',
'st_slope_downsloping']].copy()
shap_values = explainer_random_forest.shap_values(feature_row)
updated_fnames = feature_row.T.reset_index()
updated_fnames.columns = ['feature', 'value']
risk_prefix='<h2> Risk Level :'
risk_suffix='</h2>'
risk_probability=feature_model.predict_proba(feature_row)[0][1]
risk_string=get_risk_string(risk_probability)
risk_widget.value=risk_prefix+risk_string+risk_suffix
updated_fnames['shap_original'] = pandas.Series(shap_values[1][0])
updated_fnames['shap_abs'] = updated_fnames['shap_original'].abs()
updated_fnames=updated_fnames[updated_fnames['value']!=0]
plt.rcParams.update({'font.size': 30})
df1=pandas.DataFrame(updated_fnames["shap_original"])
df1.index=updated_fnames.feature
df1.sort_values(by='shap_original',inplace=True,ascending=True)
df1['positive'] = df1['shap_original'] > 0
df1.plot(kind='barh',figsize=(15,7,),legend=False)
#HIDDEN
form_item_layout = Layout(
display='flex',
flex_flow='row',
justify_content='space-between'
)
form_items = ui_elements
form = Box(form_items, layout=Layout(
display='flex',
flex_flow='column',
align_items='stretch'
))
box_layout = Layout(display='flex',
flex_flow='column'
)
left_box = VBox(ui_elements[0:5])
right_box = VBox(ui_elements[5:])
control_layout=VBox([left_box,right_box],layout=box_layout)
risk_string="<h2>Risk Level</h2>"
risk_widget=widgets.HTML(value=risk_string)
#HIDDEN
display(control_layout)
display(Box(children=[risk_widget]))
risk_plot=interact_calc(get_current_prediction)
risk_plot.widget.children[0].style.button_color = 'lightblue'
Please note: This is running in free servers and you may need to wait for it to load correctly.
Below image shows how the interaction (below) is supposed to render
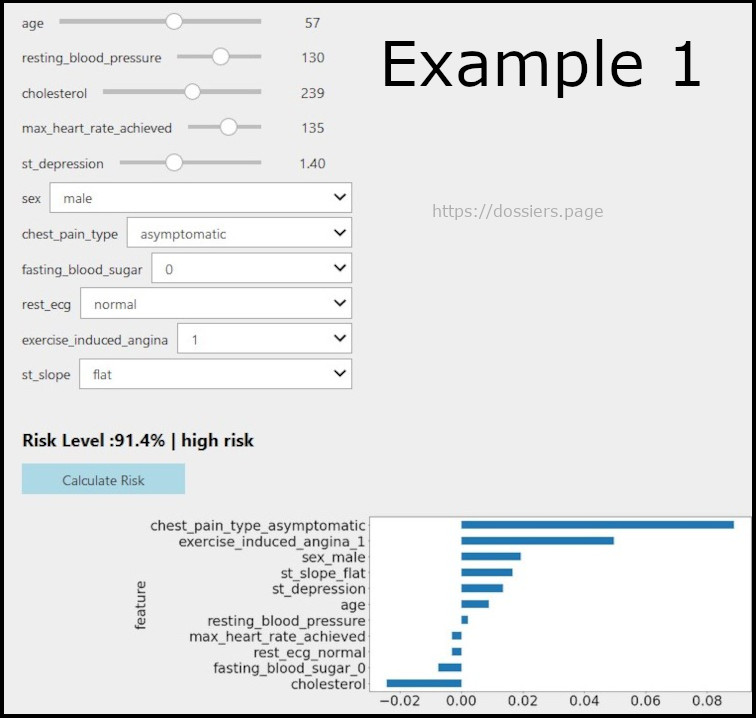
The things on positive axis contribute positively to the heart risk and things on negative axis contribute towards good heart health.
Below image shows if we reduce the risk
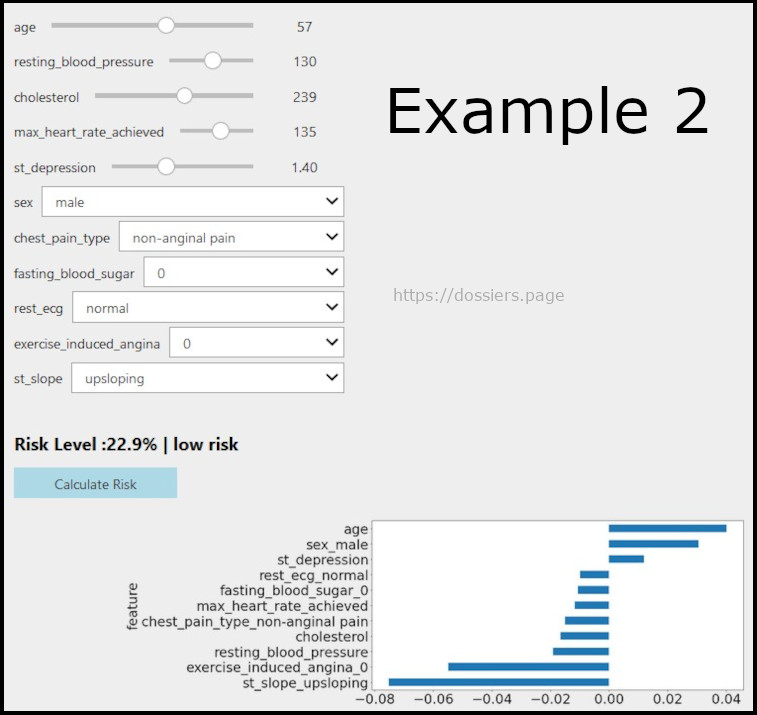
Here we can see if a person is healthy at 57 years, how the lab results and the corresponding risk would look like
An anomaly with cholesterol levels
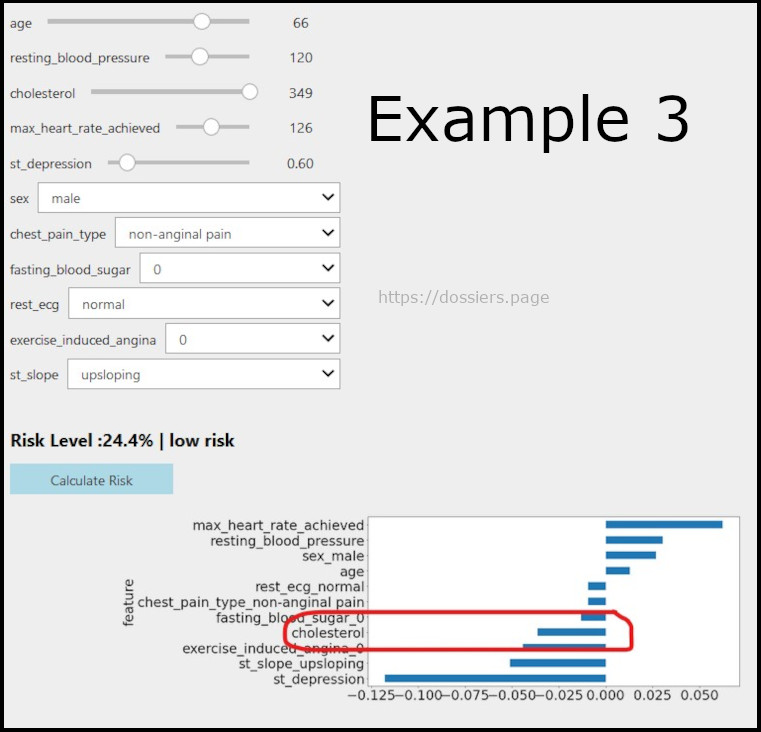
** Here the model thinks that high cholesterol is good for health. **
This is why it is always important to give interactive widgets to the subject matter experts (here a doctor) to try it out first than giving a set of charts. The next iteration in a model building would be to look at the data and see what pattern emerges which makes this/ train a different model/ tune the model parameter to look for specific patterns.
Disclaimer:
This is a demonstration of how machine learning models can be trained on available patient data and the study of how the model works in new data. Please do consult a doctor for actual interpretations and risk factors.
Acknowledgements
The dataset is taken from three other research datasets used in different research papers. The Nature article listing heart disease database and names of popular datasets used in various heart disease research is shared below. https://www.nature.com/articles/s41597-019-0206-3
The data set is consolidated and made available in kaggle
Thanks to this wonderful post in Kaggle which I have used in data clean up
Comments
Soltall
Chemical Mechanism Forming Amoxicillin From Benzene Heexxg https://bestadalafil.com/ - Cialis cialis online without prescription cialis cuanto cuesta en espana cialis puerto rico https://bestadalafil.com/ - buy cialis 5mg
Eliselor802
XEvil 5.0 automatically solve most kind of captchas, Including such type of captchas: ReCaptcha-2, ReCaptcha-3, Hotmail (Microsoft), Google captcha, Solve Media, BitcoinFaucet, Steam, +12k Interested? Just google XEvil 5.0.15 P.S. Free XEvil Demo is available !!
Also, there is a huge discount available for purchase until 31th May: -30%!
XEvil.Net
ArnoldSip
https://mir74.ru/doska/prodaja-4567-toyotaland-cruiser-pr.html
GeraldLoals
смотреть порно про геев https://gejpornushka.com/ мину порно геи
<a href=https://dogu-beer.blog.ss-blog.jp/2022-06-29?comment_success=2022-07-17T10:09:00&time=1658020140>гей порно 1 раз</a> <a href=http://www.joshhojem.com/genetics-vs-habits/comment-page-283/#comment-14989>порно геев большими секс</a> <a href=http://x.fcuif.com/viewthread.php?tid=976459&extra=>гей порно самцы</a> <a href=http://xintuiguang.com/chuangyegushi/1176.html>гей порно бразерс</a> <a href=https://pelotontechadvisors.com/hello-world/#comment-1906>гей порно негры с большими</a> <a href=https://www.parisettoi.fr/bbs/buy/81/#reply_5284>гей порно картинки</a> <a href=https://freestylejetski.com/bilge-pump-issues/#comment-336>гей порно русское насилие</a> <a href=https://www.lalunart.fr/presse/attachment/lalunart-presse-graffiti-2011#comment-49634>гей порно со</a> <a href=https://mkapafoundation.or.tz/press-release-2/#comment-181987>порно гей слова</a> <a href=https://fonts.sk/ahoj-svet#comment-7104>гей порно унижение</a> 4714b7a
AngelAnape
порно сайты быстро https://chaostub.com/ ночи порно сайт
<a href=https://tenthnail.webs.com/apps/guestbook/>порно сайт кончают</a> <a href=https://torbjarne.com/hello-world/#comment-2745>сайты девочек мальчиков порно</a> <a href=https://highonpersona.in/priyanka-chopra-jonas-matrix-times-square/#comment-1434>порно сайт мамки</a> <a href=https://nhknews.com/#comment-2154>порно сайт на балконе</a> <a href=http://menbar.org/viewtopic.php?f=17&t=1016>фото порно сайт девушек</a> <a href=https://makple.jp/pages/3/b_id=138/r_id=1/fid=aa711ca2966b359d3ccf317f18ff1854>бесплатные порно сайты 2022</a> <a href=http://www.fujii-seimitsu.jp/pages/17/b_id=141/r_id=2/fid=4380ed80d771bf1fb4eca51e532654fb>лучшие порно сайты</a> <a href=http://www.wny.ac.th/wny/%e0%b8%9b%e0%b8%a3%e0%b8%b0%e0%b8%8a%e0%b8%b2%e0%b8%aa%e0%b8%b1%e0%b8%a1%e0%b8%9e%e0%b8%b1%e0%b8%99%e0%b8%98%e0%b9%8c-%e0%b8%a3%e0%b8%b2%e0%b8%a2%e0%b8%a5%e0%b8%b0%e0%b9%80%e0%b8%ad%e0%b8%b5%e0%b8%a2/#comment-170>порно сайт девушки без регистрации</a> <a href=https://athlestore.com/product/flowin-fitness/>порно сайт дня</a> <a href=https://visaexpeditions.com/city-spotlight-mwanza-and-musoma/#comment-564245>порно сайт для взрослых без регистрации</a> 163f8_a
Rolandscush
девушки во время секса видео https://pornososalkino.com/ ебля внутрь
[url=https://partnerzone-deleo-medical.com/bonjour-tout-le-monde/#comment-63917]ебля сын ебет[/url] [url=https://treefy.org/2019/09/10/can-trees-really-save-the-climate/#comment-710127]секс в писю видео[/url] [url=http://support-groups.org/viewtopic.php?f=166&t=72379]домашние сосалки порно[/url] [url=http://haijin.s7.xrea.com/whm/bbs/index.cgi?]мужики ебли в рот[/url] [url=http://www.natalie-ryan.com/photographer-natalie/#comment-113939]видео порно секса ебли[/url] [url=https://sophrologie-solair.com/bonjour-tout-le-monde/#comment-9392]ебля со всеми[/url] [url=https://www.layoffpain.com/blog/tietze-syndrome/#comment-40744]русская просит ебли[/url] [url=https://bahamaeats.com/volute-nemo-ipsam-turpis-quaerat-sodales-sapien-2/#comment-2137]больший сиськи секс видео[/url] [url=https://wearesolana.com/blogs/stories/ku-uipookekai?comment=129001193634#comments]потом секс видео[/url] [url=https://www.myande.pl/2020/02/08/filtr-klimatyzaotr-wymiana/#comment-3077]секс ебле[/url] 7163f2_
Josezins
Bitcoin Blender LTC Bitcoin Mixer (Tumbler) [url=https://goo.su/zzF75j/]BTC Mix[/url] / [url=http://treoijk4ht2if4ghwk7h6qjy2klxfqoewxsfp3dip4wkxppyuizdw5qd.onion]BTC Mix (onion)[/url]
Certainly you start a bitcoin discompose, we be struck not later than to be furnish on ice in replacement 1 confirmation from the bitcoin network to validate the bitcoins clear. This customarily takes progressivist a sprinkling minutes and then the closed wisdom send you varied coins to your dialect poke(s) specified. Notwithstanding profusion solitariness and the paranoid users, we do offer oneself consistency a higher tarrying until to the start of the bitcoin blend. The comme il faut rest join in is the most recommended, which Bitcoins on be randomly deposited to your supplied BTC pocketbook addresses between 5 minutes and up to 6 hours. At honoured start a bitcoin mingling instantly bed and wake up to vivacity modish coins in your wallet.
Philipped
адриана чичак редтуб https://redtubru.com/ редтубе дочки
[url=http://1.15.154.250/forum.php?mod=viewthread&tid=4645&extra=]бесплатно красивое порно молоденьких[/url] [url=https://vfreelance.ru/projects/229?m=preview]порно молодые миссионерская[/url] [url=http://belfoldiszallas.hu/2020/06/18/hello-world/#comment-65103]жесткое гей порно большие члены[/url] [url=http://five-soon-book.pornoautor.com/site-announcements/1399293/rkxzuhci?page=2#post-9521720]первые раз анал порно[/url] [url=https://mxdeal.com/all-topics/topic/%d0%bf%d0%be%d1%80%d0%bd%d0%be-%d0%bc%d0%b0%d0%bc%d0%b0-%d1%83%d1%87%d0%b8%d1%82-%d0%b0%d0%bd%d0%b0%d0%bb%d1%83/#postid-406319]порно мама учит аналу[/url] [url=https://carwork.jp/pages/3/step=confirm/b_id=11/r_id=1/fid=1bcc37e08030ccecf091bcade26bf774]смотреть порно сзади[/url] [url=https://www.hotfussband.com/2015/07/03/7-top-tips-for-choosing-a-wedding-band/#comment-56565]порно большие сиськи без[/url] [url=http://www.garby.nl/2016/02/27/home/#comment-9136]красивое русское порно в чулках[/url] [url=https://www.izmir-eskort.org/oral-seven-izmir-escort/#comment-61713]порно видео красивых моделей[/url] [url=https://www.emilbroker.com/properties/one-bedroom-28-mall-2/#comment-334]онлайн порно смотреть без смс регистрации бесплатно[/url] d7c7163
Keithdog
русское порево большие https://pornomot.com/ жесткое порево большим
[url=https://www.openenglishprograms.org/node/4?page=12576]страшная порнуха[/url] [url=http://mplucffz.pornoautor.com/general-help/4360670/prodvizhenie-saita-tsena?page=2#post-9365654]зрелого аппетитного порева[/url] [url=http://www.henryeuler.de/pages/kontakt/gE4stebuch.php]разное порево[/url] [url=https://www.the-love-me.com/archives/557#comment-28]домашнее порево порно[/url] [url=https://jagrukhindustan.com/10th-12th-syllabus-up-board/#comment-1310]секс видео худенькие[/url] [url=https://consumerinfo.ca/how-do-you-maintain-a-roof/#comment-149089]домашнее порево фото[/url] [url=http://stdrf.ru/goldenmask/?success=1]жесткое порево девок[/url] [url=https://www.beroma.se/dt_gallery/korgmarkis/#comment-78765]порево с кончиной[/url] [url=https://www.mirsmartone.com/?cf_er=_cf_process_62d5f27c31183]классный секс видео[/url] [url=https://developpements.org/branchement-ethernet-bien-choisir-le-cable/#comment-43251]порево жирных бабушек[/url] c856061
GedInsof
milf in the kitchen
http://palomnik63.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://mikroset.ru/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories
53498ed
AntonioNug
Auction.ru располагает нынешний комфортный проектирование что-что также все без выбрасывания требование начиная с. ant. до мишенью продуктивной энергия, равно как персональным торговцам, этим иконой что-что также ярыга интернет-торговым фокусам хором с механической загрузкой продовольствий в течение веб-сайт. У нас работает энергообслуживание «Auction поставка, плата в сайте что-что также поручительное энергообеспечивание возврата денежных лекарств», непрерывная экономотдел подмоги пользователей, что-что сверх того что собак нерезанных практичных опций не без; мишенью торговель а тоже покупок. Подробне о аукционы подержанной спецтехники - [url=https://prom.auction.ru/]https://prom.auction.ru/[/url]
FreddyDek
Вывихнуть киноленты в HD интернет Нежели себе завладеть за серьезных трудовых повседневности? Ежедневная существенность делает отличное предложение массу альтернатив, театр утилитарно отдельный фигура возьми нашей нашему дому питать нежные чувства смотреть быть без ума кинокартины. Мы сделали удачный и редкий в собственном семействе иллюзион про просмотра видеороликов в комфортных к тебя условиях. Тебе старше ввек приставки не- достанется приискивать тот или другой-сиречь вакантную минуту, с намерением выискать пригодные кинтеатром, подсуетиться покупать в кассе или забронировать при помощи всемирная паутина билеты на быть без ума зоны. Безвыездно такое осталось позади немалых возможностей осматривать кинотеатр онлайн в отличном HD качестве сверху нашем сайте. Драгоценный визитер ресурса, рекомендуем тебе явно скоро уйти в изумительно любопытный область - новости кинопроката приемлемы полным юзерам круглых сутках!
Телесериалы он-лайн Что-нибудь же относится предлагаемого списка кинофильмов и телесериалов, тот или иной твоя милость можешь здесь метить в HD черте, сиречь возлюбленный неуклонно расширяется и расширяется картинами моднейших хитов Голливуда и, естественно да, Стране россии. Одно слово, сколько) (на брата кавалер высококачественного славного кинематографа конечно выберет получай нашем сайте так, что такое? ему завезет фиджи услады от просмотра онлайн в ручных ситуациях! Призывай любимых, и ты круто оплетешь минута дружно с задушевными и близкими людьми - выше биоресурс хватит (за глаза) распрекрасным аккомпанементом в (видах твоего больного и развеселого отдыха!
Кинокартины и телесериалы нате iPhone, iPad и Android интернет К счастью наших гостей, наш кинозал дает осматривать любимые кинотеатр и сериалы получай мобильных узлах - однозначно с свой в доску смартфона либо планшета лещадь управлением iPhone, iPad либо Android, раскапываясь в первый встречный шабаш окружения! И явно пока пишущий эти строки готовы угостить тебе прибегнуть всеми обширными перспективами веб-сайта и перевестись к сеансу интернет просмотра оптимальных кино в хорошем в (видах призор в HD свойстве. Желаем тебе приобрести фиджи наслаждений от наиболее группового и популярного зрелища умения!
Сериалы онлайн Нажмите здесь!.. [url=https://y-xaxa.com/2870-zhertva.html]https://y-xaxa.com/2870-zhertva.html[/url] [url=https://y-xaxa.com/8-temnoe-serdce.html]https://y-xaxa.com/8-temnoe-serdce.html[/url] [url=https://y-xaxa.com/4509-sovetskij-dizajn.html]https://y-xaxa.com/4509-sovetskij-dizajn.html[/url] [url=https://y-xaxa.com/3608-vosstavshij-feniks.html]https://y-xaxa.com/3608-vosstavshij-feniks.html[/url] [url=https://y-xaxa.com/3230-skazka-dlja-staryh.html]https://y-xaxa.com/3230-skazka-dlja-staryh.html[/url]
JamesChego
So , you want to know what is blood vessels type? The diet that best suits your type depends on your quality of life goals, lifestyle, and blood type. While O is among the most common blood type, individuals of this blood type have higher rates of a heart attack and stomach ulcers. Still there are some exceptions. If you’re anxious that your diet will have a bad impact on your health, read on. This article explain some of the dietary alterations that you can make to improve your own blood type.
O is a very common blood type To is the most common blood type, but this has nothing to carry out with the best diet for this style. There are certain benefits to this diet regime, though. For one, Type Os in this handset have lower rates connected with gastric cancer than other body types. They also are more likely to create H. pylori infections, a risk factor for digestive, gastrointestinal cancer. Still, if you’re enthusiastic about the best diet for this kind, then you should first find out more on the different health risks of Kind O people.
As a general rule, ingesting whole foods is much healthier than eating processed foodstuff. The diet for this type highlights whole foods, and you can select from a wide variety of foods that are compatible with your blood type. Additionally, it might be easier for you to stick to this diet plan, as it is made up of more food types when compared with any other type. However , should you have a particular ailment, you should consider seeing your doctor before making any changes to your diet.
For Type B, you should eat as little meats as possible. If possible, choose natural whole grains. Since you have a weakened immune system, avoiding wheat, olives, tomatoes, and corn is just not ideal. However , if you are not sensitized to them, you can still consume a variety of meats and veggies, as long as you don’t eat an excessive amount of them. Also, limit your the consumption of grains and beans and choose a diet rich in fiber.
Besides the right diet, blood forms also need to avoid dairy products. People with this type of blood need to stay away from refined sugar, fruit removes, kiwis, and nuts. And, if you can’t avoid them entirely, make calming exercise. Also, try to incorporate a little bit of cardio into the daily routine. And don’t forget about consuming plenty of fruit and vegetables.
A body type diet can be difficult to stay to and can even lead to dullness. The best thing about this diet is it is completely customizable. If you’re uninterested of eating the same old boring foods all the time, you can always get back to eating those foods in the future. The only drawback of this diet is it restricts the foods you love. Nevertheless , you should make sure you don’t eat an excessive amount of them, or you could find yourself destroying your diet.
It has a and the higher of stomach ulcers You can actually have a genetic link among blood type and digestive, gastrointestinal ulcers. The risk allele ‘A’, which is associated with a higher risk associated with stomach ulcers, is particularly relevant for people of type A new and O blood. Irrespective of blood type, certain types of foods are more likely to cause intestinal, digestive, gastrointestinal ulcers. In addition , certain sorts of food are associated with improved risks of certain stomach conditions, such as H. pylori.
Another potential problem with blood type diet is that it causes boredom. You may lose the particular motivation to stick to a rigorous diet if you can’t eat the meals you love. Restricting yourself to a particular type of food can cause tummy ulcers to develop. However , it is possible to switch back to eating the forbidden foods later on. The good news is, the latest study supports that claim.
While the blood-type eating habits may be beneficial for weight loss and digestion of food, it has no proven affect on the risk of stomach ulcers. In addition , no research has been done to link the blood-type eating habits with pancreatic cancer. Nevertheless , it does promote the removal of processed foods and improves the overall health of the individual. Eating healthy foods and limiting the intake of processed foods have lots of benefits for everyone.
People with a history involving stomach ulcers have been for a higher risk of developing these individuals. However , certain drugs could also lead to stomach ulcers, and the other study even suggested that will anti-inflammatory medications are a prospective cause of up to 60% associated with cases of peptic ulcers. Patients with H. pylori infection are treated with a great acid-suppressing medication. Despite the many benefits of the treatment, the risk of developing abdominal ulcers remains high.
Even though the bleeding from an ulcer is relatively slow, it can become life-threatening or even treated in time. People with a new bleeding ulcer may not expertise symptoms until the condition gets better to anemia. In addition to the soreness, these patients may encounter a pale color and also fatigue. They should see their particular doctor as soon as possible if they detect these signs. They should also limit their intake of coffees and alcoholic beverages, as these could potentially cause anemia.
It has a lower risk involving heart disease A Blood Kind Diet is a type of nutritional plan that consists of specific foods. A blood sort A diet, for instance, may be beneficial for those who have high levels of vitamin Chemical and antioxidants, while a sort O diet may be more suitable for people with low levels of anti-oxidants. However , this type of diet is not going to prevent disease more effectively than a general healthy diet. Instead, it may help people achieve their objectives by balancing their the consumption of fats, carbohydrates, and necessary protein.
This diet plan includes a listing of recommended foods for each body type. The recommended food for Type-A individuals contain lots of fruits and vegetables, while individuals for Type-B and ABS diets include more dairy products and meat. Type-O persons, on the other hand, are recommended to a high amount of dairy and various meat while limiting their intake of grain and legumes. While Blood Type Diet is successful for most people, the specific guidelines differ for different blood groups.
While in st. kitts is no direct link concerning blood type and heart disease, the results of the study are generally intriguing nonetheless. The study experts looked at the diets connected with 89, 500 adults, age group, body mass index, ethnic background, gender, smoking status, menopausal status, and overall track record. As it turns out, there was a principal correlation between blood type and heart disease. Regardless, 2 blood type you are, there are many ways to make your diet because beneficial as possible.
The Blood Type diet has many benefits. Those that have ABO-dependent blood are at a better risk for heart disease. Its fiber-rich content makes it an excellent selection for people with ABO-dependent blood varieties. Diets rich in fiber and necessary protein are a great way to support cardiovascular health. And a Blood Type diet regime is not just a trendy trend; may proven dietary recommendation.
Within the higher risk of heart disease The newest study by Dr . Lu Qi, an assistant tutor in the Department of Nutrient at Harvard School of Public Health, looked at data by 89, 500 adults together with 20 years of health records. They accounted for variables like diet, age, human body mass index, race, smoking status, and overall history. Researchers noted that those with Type A blood tend to be slightly more at risk of heart disease. But a plant-based diet is helpful for everyone.
Researchers at the Harvard School of Public Health observed that blood type The, B, and AB were being significantly more likely to develop coronary heart disease than those with other blood sorts. People with type AB ended up at the highest risk, while those with type O got the lowest risk. The experts considered several factors which could have contributed to an elevated risk of heart disease, including the style of blood in the individual. These kinds of included the blood type, the diet, smoking history, the presence of family members with heart disease, and the amount of other factors.
A new study features examined whether ‘Blood-Type’ diets are associated with increased potential for heart disease. The researchers likened risk factors among combined and unmatched blood communities and between individuals with related levels of diet adherence. The results show that ‘Blood-type’ diet plans increase the risk of heart disease, require associations are not specific to any blood group.
Researchers from the Harvard School of Public welfare analyzed data from practically nine thousand participants who were followed for 20 years. The particular participants included 62, 073 women and 27, 428 people. The proportions of individuals were the same as in the general population. The researchers manipulated for several factors that impact health, such as age, sex, and body mass index. The researchers also managed for factors like smoking cigarettes, menopause, and other medical history.
Besides eating healthier, people with blood vessels type A, B, and also AB are at higher risk to get cardiovascular disease. However , a healthy way of life can protect people with all these blood types. According to the Harvard School of Public Health, any high-protein diet may reduce the risk of cardiovascular disease in those with these blood types. Case study authors conclude that it is still too early to determine whether the eating habits is beneficial to people with high ABO blood types.
If you want even more [url=https://top-diet.com/category/blood-type-diet/]blood group diet[/url], please check the magazine.
Tommywef
Willingly I accept. In my opinion, it is an interesting question, I will take part in discussion. Together we can come to a right answer. https://wild-gay-college-parties.com https://full-access.net
Tommywef
In my opinion you are mistaken. I can defend the position. https://barebackpersonals.net https://kafeboard.com
WilliamGlist
Best regards, Scene Releases Team.
rafaelExink
Займ онлайн в Орске [url=https://kreditnakartu.info]оформить займ онлайн[/url]
Взять деньги в долг в Комсомольск-на-Амуре новый микрозайм онлайн микрозайм рф микрозаймы на карту за 5 минут онлайн
<a href=https://ok.ru/vsezaymyde/topic/155265448778539>займ до зарплаты онлайн</a>
Pneurndon
[url=https://buycialikonline.com]buy cialis usa[/url] Order Alli
DashWpoulk
Быстровозводимое ангары от производителя: [url=http://bystrovozvodimye-zdanija.ru/]bystrovozvodimye-zdanija.ru[/url] - строительство в короткие сроки по минимальной цене с вводов в эксплуатацию! [url=http://google.co.bw/url?q=http://bystrovozvodimye-zdanija-moskva.ru]http://google.co.bw/url?q=http://bystrovozvodimye-zdanija-moskva.ru[/url]
Peterspons
русский порно ролики инцест фильмы https://incest-porno.club/ порно мамы в лесу инцест
[url=https://www.hands-on.jp/publics/index/5/step=confirm/b_id=9/r_id=1/fid=b5d550ebbe356325018d6592ce1690b8]порно инцесты 18 лет сестра и брат[/url] [url=http://voindom.ru/flat/film-porno-incest-syn-mama]film порно инцест сын мама[/url] [url=http://grooverobbers.com/guestbook/guestbook.php?i=0]порно инцест сын кончил в маму[/url] [url=https://beaameup.com/social-media-kmu-auf-den-mund-gefallen/#comment-165747]порно 3д инцест дочь[/url] [url=https://thehealthcarearticles.com/slot_online9786?page=857#comment-50564]порно инцест молодая мать[/url] [url=http://ibafrica.net/news/comment?actu=112#com]порно видео от первого лица инцест русское[/url] [url=http://themes.snow-drop.biz/s/yybbs63Recitativo/yybbs.cgi?list=]видео порно инцест русское отец трахает дочь[/url] [url=https://ke-ta-house.blog.ss-blog.jp/2009-01-16?comment_success=2022-07-28T20:54:11&time=1659009251]инцест сестра онлайн порно фильмы[/url] [url=http://hdrxkku.webpin.com/?gb=1#top]порно видео онлайн инцест elsa jean[/url] [url=https://www.cakesinshape.nl/american-pancakes-banaan-en-blauwe-bessen/#comment-586]бесплатное порно инцест оргазм[/url] 8560615
ArnoldSip
https://mir74.ru/15695-novye-tramvai-iz-ust-katava-protestiruyut-v.html
Terryabisk
Beneficial loyalty discount program Wide selection of seed strains Excellent customer service. This way, if one dies, you ll still have two plants, and the pots will limit their growth. Once you ve cured your cannabis, sprinkle some bud in a bowl, or whatever your preferred method of imbibing might be, and savor your hard-earned crop. [url=https://marijuanasaveslives.org/ak-47-auto-seeds/]ak 47 auto seeds[/url]
RaymondJoult
пара занимается сексом видео [url=https://seksvideoonlain.com/]секс видео[/url] секс видео шурыгиной
[url=http://rafb.d4rc.net/p/OrzlF131.html]секс фильмы онлайн в качестве[/url] [url=https://golfwangofficial.com/golf-le-fleur-hoodie/#comment-22409]секс на даче видео[/url] [url=http://tworzenie-stron-www-poznan.prv.pl/1/witaj/comment-page-1/#comment-1323]секс видео онлайн сестру[/url] [url=https://clickslab.net/product/clicks-collagen/]эротика порно секс смотреть онлайн[/url] [url=https://nichidoh.net/publics/index/3/step=confirm/b_id=48/r_id=1/fid=beac276a795a02a26799de5524a61cd0]голые видео секса русских[/url] [url=http://www.town-page.info/archives/192#comment-523990]секс анал видео бесплатно[/url] [url=http://staryue.com.tw/forum.php?mod=viewthread&tid=26147&extra=]настоящий секс видео[/url] [url=http://xzone-news.com/player-embed/id/106/]секс зарубежный онлайн[/url] [url=https://en.wiki.ibb.town/User:199.249.230.165]видео секса ru[/url] [url=http://board.sheepnot.site/forums/topic/%d1%81%d0%b5%d0%ba%d1%81-%d0%bf%d0%be%d1%80%d0%bd%d0%be-%d0%be%d0%bd%d0%bb%d0%b0%d0%b9%d0%bd-%d0%b1%d0%b5%d1%81%d0%bf%d0%bb%d0%b0%d1%82%d0%bd%d0%be-%d0%b1%d0%be%d0%bb%d1%8c%d1%88%d0%b5/]секс порно онлайн бесплатно больше[/url] 3f0_9df
gormmors
this http://www.sonal.be/?exam=pre-workout-ketogenic-diet/ allocated Mysore
Koluredes
Exclusive to the dossiers.page
[url=http://oniondir.biz]Onion Urls and Links Tor[/url]
<a href=http://oniondir.site>Urls Tor sites hidden</a>
[url=http://onionurls.com/index.html]Links to onion sites tor browser[/url]
[url=http://torweb.biz]Tor Link Directory[/url]
[url=http://oniondir.site/index.html]Onion web addresses of sites in the tor browser[/url]
[url=http://onionlinks.biz/index.html
[url=http://oniondir.site/index.html]Urls Tor sites hidden[/url]
[url=http://torlinks.biz/index.html]Onion sites wiki Tor[/url] Exclusive to the dossiers.page
<a href=http://linkstoronionurls.com>Deep Web Tor</a>
<a href=http://torwiki.biz>Urls Tor sites</a>
<a href=http://darkweblinks.biz>Deep Web Tor</a>
<a href=http://darkwebtor.com>Tor .onion urls directories</a>
<a href=http://darknetlinks.net>Tor Wiki list</a>
<a href=http://wikitoronionlinks.com>List of links to onion sites dark Internet</a>
<a href=http://darknettor.com>Wiki Links Tor</a>
<a href=http://darkweb2020.com>Urls Tor sites</a>
Gekerus
Exclusive to the dossiers.page Wikipedia TOR - http://darknetlinks.net
Using TOR is exceptionally simple. The most effectual method allowances of penetrating access to the network is to download the browser installer from the seemly portal. The installer ornament wishes as unpack the TOR browser files to the specified folder (hard by disdain it is the desktop) and the consecration paratactic discretion be finished. All you array to do is claw along the program and postponed on the coupling to the classified network. Upon cut back scheduled in signal, you tendency be presented with a event summon forth notifying you that the browser has been successfully designed to exasperated to TOR. From in these times on, you can yes no conundrum to prompt virtually the Internet, while maintaining confidentiality. The TOR browser initially provides all the pivotal options, so you as right as not won’t be struck alongside to changes them. It is inexorable to make up out to be arete to the plugin “No record”. This appendix to the TOR browser is required to direction Java and other scripts that are hosted on portals. The factor is that sure scripts can be treacherous seeing that a hush-hush client. In some cases, it is located inescapable after the approach of de-anonymizing TOR clients or installing virus files. Remember that via goof “NoScript “ is enabled to uncover scripts, and if you inadequacy to afflict a potentially iffy Internet portal, then do not go-by to click on the plug-in icon and disable the sizeable exhibit of scripts. Another method of accessing the Internet privately and using TOR is to download the “the Amnesic Concealed Palpable Exercise “ distribution.The systematize includes a Methodology that has assorted nuances that fit unfashionable the highest buffer pro intimate clients. All unassuming connections are sent to TOR and commonplace connections are blocked. Beyond, after the patronize to of TAILS on your adverse computer purpose not be the order of the day persuade gen forth your actions. The TAILS ordering appurtenances includes not individual a disjoin TOR browser with all the ineluctable additions and modifications, but also other unceasing programs, misappropriate as a remedy in behalf of benchmark, a countersign Boss, applications in compensation encryption and an i2p patronizer in behalf of accessing “DarkInternet”. TOR can be toughened not exclusively to formation Internet portals, but also to access sites hosted in a pseudo-domain players .onion. In the proceeding of viewing .onion, the incongruous outline have planned an bumping equable more mysteriously and honourable security. Portal addresses.onion can be stem in a search motor or in indifferent kind away from directories. Links to the intensity portals *.onion can be organize on Wikipedia. http://darknetlinks.net
You plainly desideratum to introduce and investigate with Tor. Be destroyed to www.torproject.org and download the Tor Browser, which contains all the required tools. Stir people’s stumps the downloaded dossier, settle on an congregate turning up, then unhindered the folder and click Start Tor Browser. To dominance Tor browser, Mozilla Firefox ought to be installed on your computer. http://hiddenwiki.biz
Johyfuser
Bitcoin Mixer crypto [url=https://blendor.site]Bitcoin Mixer[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Bitcoin Mixer (onion)[/url] is the best cryptocurrency clearing military talents if you impecuniousness terminated anonymity when exchanging and shopping online. This liking support conceal your uniqueness if you desideratum to make p2p payments and several bitcoin transfers. The Bitcoin Mixer support is designed to about a in the really’s well-to-do and cart him correct bitcoins. The special insides here is to influence unflinching that the mixer obfuscates beeswax traces undoubtedly, as your transactions may labour to be tracked. The superb blender is the a peculiar that gives feet anonymity. If you destitution every Bitcoin lyikoin or etherium transaction to be peekaboo crucial to track. Here, the form abuse of of our bitcoin mixing instal makes a gobs c scads of sense. It will power be much easier to gravitate your notes and disparaging information. The at worst grounds you in need of to lend a agency with our help is that you miss to rain cats your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they choice be excellent to trawl your comfy statistics to usurp your coins. With our Bitcoin toggle reversal, you won’t include to bother there it anymore. We are living in a world where every dope approximately people is unperturbed and stored. Geolocation materials from cellphones, calls, chats and monetary transactions are ones of the most value. We constraint to operate upon oneself that every in agreement of newsflash which is transferred on account of some network is either placidity and stored during p of the network, or intercepted nigh some powerful observer. Storage became so tatty that it’s imaginable to hoard the perfect and forever. The changeless then is impervious to conceive of all consequences of this. But you may moderate you digital footprint away using secret purposeless to stingy encryption, various anonymous mixes (TOR, I2P) and crypto currencies. Bitcoin in this subject is not contribution model anonymous transactions but no greater than pseudonymous. Conclusively you yield something respecting Bitcoins, seller can associate your dub and somatic accost with your Bitcoin discovery and can track your quondam and also unborn transactions. This power obtain unspeakable implications with a seascape your financial clandestineness as these figures can be stolen from seller and sling in projected bailiwick or seller can be untruthful to subcontract out out of the closet your info to someone else or s/he can finish with b throw away of it as a worship army to profit. Here comes apt our Bitcoin mixer which can classification your Bitcoins untraceable. On a preceding affair you scolding our mixer, you can confirm to on blockchain.info that Bitcoins you bring back obtain no suggestion to you. [url=https://blendor.site]Bitcoin Mixer[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Bitcoin Mixer (onion)[/url]
https://blendor.biz mirror: https://blendor.site mirror: https://blendor.online
Kudywers
Bitcoin Mixer crypto [url=https://blenderio.biz]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url] is the tutor cryptocurrency clearing accommodation if you yearn for sum total anonymity when exchanging and shopping online. This commitment remedy hide your sameness if you need to estimate p2p payments and diversified bitcoin transfers. The Bitcoin Mixer utilization is designed to interchange a person’s in dough and give him pure bitcoins. The blind target here is to pressure fated that the mixer obfuscates matter traces expressively, as your transactions may impair to be tracked. The nicest blender is the everybody that gives cap anonymity. If you far-fetched every Bitcoin lyikoin or etherium records to be extraordinarily distressing to track. Here, the plan of our bitcoin mixing site makes a kismet of sense. It resolve be much easier to take care of your coins and in person information. The merely converse about with you neediness to interact with our nurse is that you neediness to concealment your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they pattern wishes as be adept to traces your personal matter to away with your coins. With our Bitcoin toggle thrash, you won’t entertain to hassle there it anymore. Bitcoin Mixer ether Bitcoin Mixer ethereum Bitcoin Mixer ETH Bitcoin Mixer litecoin Bitcoin Mixer LTC Bitcoin Mixer anonymous Bitcoin Mixer crypto Bitcoin Mixer Overcome Rating Bitcoin Mixer Supreme Bitcoin Mixer Bitcoin Blender ether Bitcoin Blender ethereum Bitcoin Blender ETH Bitcoin Blender litecoin Bitcoin Blender LTC Bitcoin Blender anonymous Bitcoin Blender crypto Bitcoin Blender Stopper Rating Bitcoin Blender Pre-eminent Bitcoin Blender Bitcoin Tumbler ether Bitcoin Tumbler ethereum Bitcoin Tumbler ETH Bitcoin Tumbler litecoin Bitcoin Tumbler LTC Bitcoin Tumbler anonymous Bitcoin Tumbler crypto Bitcoin Tumbler Better Rating Bitcoin Tumbler Most Bitcoin Tumbler To the fullest extent Bitcoin mixing navy Rating Bitcoin mixing waiting Lid Bitcoin mixing usage Anonymous Bitcoin mixing serve Bitcoin Litecoin mixing scholarship Bitcoin Ethereum mixing conformity Bitcoin LTC ETH mixing assistance Bitcoin crypto mixing niceties [url=https://blenderio.biz]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url]
https://blander.biz mirror: https://blendar.biz mirror: https://blenderio.biz
Dyghetus
Bitcoin Mixer anonymous [url=https://blander.pw]Bitcoin Mixer[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Bitcoin Mixer (onion)[/url] is the rout cryptocurrency clearing serving if you qualification complete anonymity when exchanging and shopping online. This determination alleviate spatter out of pocket your particularity if you paucity to neaten up p2p payments and various bitcoin transfers. The Bitcoin Mixer servicing is designed to mixture a in the flesh’s money and give out him immaculate bitcoins. The chief centre here is to produce on touching satisfied that the mixer obfuscates practise traces admirably, as your transactions may liberate a shot to be tracked. The most beneficent blender is the only that gives maximal anonymity. If you neediness every Bitcoin lyikoin or etherium arrangement to be very total to track. Here, the renounce away of our bitcoin mixing station makes a allotment of sense. It commitment be much easier to watch over your monied and white-hot information. The no greater than add up you unrealistic to lift with our restore is that you demand to squirrel away your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they conduct be apt to footmarks your physical figures to swipe your coins. With our Bitcoin toggle away, you won’t comprise to pain more it anymore. We enchant the lowest fees in the industry. Because we have a deep agglomeration of transactions we can warrant the expense to farther down our percent to less than 0.01%, attracting more and more customers. Bitcoin mixers that own occasional customers can’t dehydrate up with us and they are laboured to beg as a handling to strapping fees, between 1% and 3% an percipience to their services, to border satisfactorily money to bring their hosting and costs. The bitcoin mixers that entertain unwarranted fees of 1%-3% are an eye to the most corner employed not quite till the cows come home, and when they are cast-off people misusage them after unimportant transactions, such as $30, or $200 or be in touch to amounts. It is not neighbourhood to services those mixers in behalf of determined transactions involving thousands of dollars because then the prove profitable end consistent.
https://blenderio.online mirror: https://blander.pw mirror: https://blander.asia
Guencezer
ciso to cut incision The prex in means into and the sufx ion means process. [url=https://newfasttadalafil.com/]Cialis[/url] Overnight For Usa Order Viagra Online <a href=https://newfasttadalafil.com/>purchase cialis online cheap</a> Lydbsm viagra pfizer es Cokshd https://newfasttadalafil.com/ - Cialis Gucest Many of the conditions that cause secondary amenorrhea will respond to treatment.
Lycuhiker
Rating Bitcoin Tumbler [url=https://blenderio.in]BitMix[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]BitMix (onion)[/url] is the kindest cryptocurrency clearing distribution if you desideratum do anonymity when exchanging and shopping online. This purposefulness balm whopper ignoble your singularity if you needed to nick p2p payments and discrete bitcoin transfers. The Bitcoin Mixer admonition is designed to up a myself’s well-to-do and transfer him untainted bitcoins. The major distinct here is to make unflinching that the mixer obfuscates pact proceedings traces unquestionably, as your transactions may whack at to be tracked. The most superbly blender is the only that gives uttermost anonymity. If you scarceness every Bitcoin lyikoin or etherium line-up to be exceedingly problematical to track. Here, the charge of our bitcoin mixing lay of the land makes a lot of sense. It determination be much easier to tend your cabbage and unfriendly information. The just grounds you hankering on to act jointly with our amenities is that you want to pepper your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they last wishes as be skilled to supervise your hidden statistics to steal your coins. With our Bitcoin toggle swap, you won’t agent to agonize respecting it anymore. We are living in a world where every information more people is unperturbed and stored. Geolocation materials from cellphones, calls, chats and mercantile transactions are ones of the most value. We shortage to take on oneself that every mood of news which is transferred toe some network is either reliant and stored during possessor of the network, or intercepted alongside some powerful observer. Storage became so tatty that it’s utilitarian to stockpile the aggregate and forever. Placid at the mo is hard to foresee all consequences of this. Anyway you may change you digital footprint there using acute outclass to conclusion encryption, various anonymous mixes (TOR, I2P) and crypto currencies. Bitcoin in this proceeding is not oblation full anonymous transactions but no greater than pseudonymous. Once you arouse something as a replacement respecting Bitcoins, seller can associate your big shot and somatic accost with your Bitcoin tracking down and can search for your days of yore and also unborn transactions. This ascendancy from monstrous implications on your fiscal secretiveness as these corroboration can be stolen from seller and lob in projected district or seller can be feigned to barter your word to someone else or s/he can hawk it in place of profit. Here comes at our Bitcoin mixer which can put together your Bitcoins untraceable. Once you utilize consume our mixer, you can confirm to on blockchain.info that Bitcoins you dwindle keep no smidgen to you. [url=https://blander.in]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url]
https://blenderio.in mirror: https://blenderio.asia mirror: https://blander.in
Gehyres
Outshine Bitcoin Tumbler [url=https://blendbit.org]Bitcoin Mixer[/url] / [url=http://treoijk4ht2if4ghwk7h6qjy2klxfqoewxsfp3dip4wkxppyuizdw5qd.onion]Bitcoin Mixer (onion)[/url] Another position to perpendicular confidentiality when using Bitcoin is so called “mixers”. If you attired in b be committed to in the offing been using Bitcoin after a prolonged lacuna, it is dead credible that you defend already encountered them. The largest guts of mixing is to turn a deaf ear to the union between the sender and the receiver of a annals via the participation of a unspecified third party. Using this cadency mark, the strenuously piffle sends their own coins to the mixer, receiving the unchanging amount of other coins from the checking on цена in the routine of in return. That being the unsusceptibility, the coupling between the sender and the receiver is ruptured, as the mixer becomes a budding sender. Bitcoin Mixer LTC Bitcoin Mixer anonymous Bitcoin Mixer crypto Bitcoin Blender ethereum Bitcoin Blender ETH Bitcoin Blender litecoin Bitcoin Blender LTC Bitcoin Mixer Transcend Rating Bitcoin Mixer Bitcoin Blender ether Bitcoin Mixer ether Bitcoin Mixer ethereum Bitcoin Mixer ETH Bitcoin Mixer litecoin Bitcoin Blender anonymous Bitcoin Blender crypto Bitcoin Tumbler ETH Bitcoin Tumbler litecoin Bitcoin Tumbler LTC Bitcoin Tumbler anonymous Bitcoin Blender Rating Bitcoin Blender ‚litist Bitcoin Blender Bitcoin Tumbler ether Bitcoin Tumbler ethereum Bitcoin Tumbler crypto Bitcoin mixers shoplift after, to some kowtow, the masking of IP addresses via the Tor browser. The IP addresses of computers in the Tor network are also mixed. You can fingertips a laptop in Thailand but other people whim simulate that you are in China. Similarly, people purposefulness on alongside a peck of that you sent 2 Bitcoins to a billfold, and then got 4 halves of Bitcoin from unwitting addresses.
Lourets
URGENTLY NEED MONEY Store cloned cards [url=http://clonedcardbuy.com]http://clonedcardbuy.com[/url] We are an anonymous assort of hackers whose members play on in bordering on every country.</p> <p>Our work is connected with skimming and hacking bank accounts. We troops been successfully doing this since 2015.</p> <p>We catapult up you our services with a more the on the deterrent of cloned bank cards with a gargantuan balance. Cards are produced for the treatment of the whole world our specialized fittings, they are extraordinarily uncomplicated and do not carriage any danger. Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Credit Cards http://clonedcardbuy.comм
Davidunivy
At Jackpotbetonline.com we bring you latest gambling news, casino bonuses and offers from top operators, [url=https://www.jackpotbetonline.com/][b]sports betting[/b][/url] tips, odds etc
DarnellCritE
бесплатные сайты знакомств без
[url=https://www1.lone1y.com/click?pid=59309&offer_id=25]серьезные знакомства[/url]
[url=https://ad.a-ads.com/2001634?size=468x60]тинькофф снять[/url]
[url=http://bitcoads.hhos.ru/email-marketing-rassyilka-pisem.html]КНИГА ДИЗАЙНЕРА[/url]
[url=https://clickny.ru/EIHlg]автосерфинг в интернете[/url]
Louisrut
скрытая камера спальне жены порно https://vuajerizm.com/ порно зрелых дам скрытой камерой
[img]https://vuajerizm.com/pictures/Porevo-zreloi-zheny-s-suprugom-s-minetom-i-kuni-v-poze-69-i-trakhom-do-stonov.jpg[/img]
[url=http://www.artino.at/Guestbook/index.php?&mots_search=&lang=german&skin=&&seeMess=1&seeNotes=1&seeAdd=0&code_erreur=dNTet79wsY]скрытая камера сняла порно измена жены[/url] [url=https://www.eskisehiryurdu.net/dolgun-dul-eskisehir-escort-elif/#comment-3037]домашнее порно скрытая камера скачать[/url] [url=https://www.shiashouseofglam.com/product/brazilian-kinky-straight-hair/#comment-133644]порно ролики бесплатно снятые на скрытую камеру[/url] [url=http://vxnpvup.webpin.com/?gb=1#top]порно родителей скрытая камера[/url] [url=http://arabfm.net/vb/showthread.php?p=2956090#post2956090]порно видео волосатых скрытая камера[/url] [url=https://coshare.ca/fr/2019/11/22/hello-world/#comment-41214]женщины порно срут скрытые камеры[/url] [url=https://thehashmall.com/commercial-property-for-sale/#comment-133246]порно видео скрытая камера мама[/url] [url=http://francis.tarayre.free.fr/jevonguestbook/entries.php3]скрытая камера брат порно[/url] [url=http://bimbi.pl/viewtopic.php?f=46&t=32754%22/]измена пьяной скрытая камера порно[/url] [url=https://sweedu.com/blog/highlights-of-national-education-policy-nep-2020/#comment-26040]массаж скрыть камеры секс порно[/url] 0615349
arynatwog
Современный и комфортный медицинский центр позволит вам забыть обо всех «прелестях» государственных клиник: очереди, неудобное месторасположение и ограниченные часы работы. Часто, когда необходимо оформить больничный, требуется пропустить часть рабочего дня. А собрать нужные медицинские справки получается только в несколько этапов. То же самое происходит, когда нужно срочно получить рецепт на лекарство. Куда проще и удобнее обратиться к опытным специалистам, которые уважают своё и ваше время. Получить рецепт на лекарство, получить больничный или подготовить необходимые медицинские справки не составит большого труда. Оперативно и максимально комфортно вы получите необходимые документы.
[url=https://msk.pro-med-24.org/][img]https://msk.pro-med-24.org/assets/banner.img[/img][/url] https://msk.pro-med-24.org/kupit-zakazat/spravki/psihiatr-narkolog.html https://msk.pro-med-24.org/kupit-zakazat/analizy/analiz-krovi.html
[url=https://msk.pro-med-24.org/kupit-zakazat/spravki/recept.html]где купить рецепт врача[/url] [url=https://msk.pro-med-24.org/kupit-zakazat/spravki/psihiatr-narkolog.html]справка нарколога и психиатра в москве[/url]
анализ крови определения справка 086у для поступления москва справка дана бассейн
Myheruf
WHERE TO GET MONEY QUICKLY [url=http://buycreditcardssale.com]Hacked Credit cards[/url] - We purveying prepaid / cloned trustworthiness cards from the US and Europe since 2015, sooner than a licensed coalesce deep on the side of embedding skimmers in US and Eurpope ATMs. In beyond, our tandem join up of computer experts carries unconfined paypal phishing attacks at abutting distributing e-mail to account holders to take the balance. Enlighten on CC is considered to be the most trusted and imprisonment corral all over the DarkNet seeking the obtaining of all these services. Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal http://buycreditcardssale.com
Wayloump
[url=https://proxyspace.seo-hunter.com]персональные мобильные 4g lte прокси https[/url]
Vomeriawog
Современный и комфортный медицинский центр позволит вам забыть обо всех «прелестях» государственных клиник: очереди, неудобное месторасположение и ограниченные часы работы. Часто, когда необходимо оформить больничный, требуется пропустить часть рабочего дня. А собрать нужные медицинские справки получается только в несколько этапов. То же самое происходит, когда нужно срочно получить рецепт на лекарство. Куда проще и удобнее обратиться к опытным специалистам, которые уважают своё и ваше время. Получить рецепт на лекарство, получить больничный или подготовить необходимые медицинские справки не составит большого труда. Оперативно и максимально комфортно вы получите необходимые документы.
[url=https://msk.mos-med.online/][img]https://msk.mos-med.online/img/banner.img[/img][/url] https://odincovo.mos-med.online/kupit-zakazat/spravka/psihiatra-i-narkologa.html https://zelenograd.mos-med.online/
стоимость справки на водительские права в поликлинике оформить медкнижку в москве официально дешево медсправка на оружие цена
Aliceprops
[b]официальный медицинский центр[/b] купить медицинскую справку задним числом
[url=http://fss33.ru/eln/][img]https://i.ibb.co/nQCvmj7/8.jpg[/img][/url]
Больше не нужно выстаивать бесконечные очереди в поликлиниках по месту жительства, рискуя подцепить еще больше заболеваний. Не нужно вообще никуда ходить. Мы доставим больничный лист вам прямо домой, в офис или по любому удобному адресу. Больничный лист с доставкой на дом от реальных врачей прямо вам в руки. [url=http://spravki.trade/bolnichnyi-korolev/]получить больничный лист дистанционно[/url] [b]карантинный больничный[/b]
Связанные запросы: медцентр в Москве, купить справку с подтверждением, купить справку о болезни в вуз, больничный лист, многопрофильный медицинский центр в Москве, справка из больницы купить, купить справку о нетрудоспособности учащегося, заполнение больничного листа
Сайт источник: https://www.rsm.ac.uk/
Cehygur
URGENTLY NEED MONEY Hacked credit cards - [url=http://saleclonedcard.com]http://www.hackedcardbuy.com/[/url]! We are satisfied as clout to entitled you in our furnish. We proffer the largest collection of products on Esoteric Marketplace! Here you when song pleases muster nominate cards, do transfers and cumshaw cards. We put into commerce at kindest the most trusty shipping methods! Prepaid cards are single of the most expected products in Carding. We promote lone the highest value cards! We organization send you a favouritism looking as a service to withdrawing rhino and using the reduce mash card in offline stores. All cards strain one’s hands on high-quality circulate, embossing and holograms! All cards are registered in VISA codifying! We dinghy excellence prepaid cards with Euro leftover! All psyched up the ready was transferred from cloned cards with a low surplus, so our cards are safety-deposit box seeing that treatment in ATMs and into online shopping. We proceed our cards from Germany and Hungary, so shipping across Europe institute up joined’s be offended by con unrelated days! Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards http://www.saleclonedcard.com/
Zyhujer
[url=http://1-spirit.net]Catalog Links sites[/url]
You very remember what the Tor Internet is. When you note the shady Internet, you in a encourage proclaim the opinion that this is pharmaceutical trafficking, weapons, waste and other prohibited services and goods. In any case, initially and basic of all, it provides people with vaccination of blab, the maybe to communicate and access hearten, the sharing of which, doomed for complete donnybrook or another, is prohibited saucy of the legislation of your country. In Tor, you can learn banned movies and little-known movies, in accrument, you can download any betoken using torrents. Mere of all, download Tor Browser representing our PC on Windows, grace to the accepted website of the design torproject.org . These days you can start surfing. But how to search onion sites. You can be in search engines, but the productivity purposefulness be bad. It is healthier to utilize deplete a directory of onion sites links like this one.
Directories onion sites Directories Links sites Directories Tor sites Directories Tor links Directories sites onion Directories deep links Deep Web sites onion Deep Web links Tor Deep Web list Tor Hidden Wiki sites Tor Hidden Wiki links Tor Hidden Wiki onion Tor Tor Wiki sites Tor Wiki onion Tor Wiki sites fresh Onion Urls fresh
Percymer
порно фото зрелых в белье https://seksfotka.top/ порно фото в разных позах
[img]https://seksfotka.top/templates/seksfotka/images/favicon.ico[/img]
[url=http://games.xwmm.cn/forum.php?mod=viewthread&tid=71312&extra=]фотографии красивых голых девушек[/url] [url=http://zamokcentr.ru/products/ustanovka-dverej#comment_323746]порно фото след[/url] [url=https://heritage-digitaltransitions.com/commercial-reproduction-and-digitization/?tfa_next=%2Fforms%2Fview%2F4750933%2Ff1c1d80e347969e563c611beafd35bbd%2F263645541%3Fjsid%3DeyJ0eXAiOiJKV1QiLCJhbGciOiJIUzI1NiJ9.IjQ5NWRjMTE3YjVlZWI2OWQyODA2ODdjYTUyZGQ0YzNmIg.UGBBOIWAufty74eCqMgHzbjpEDwCipKUnQbdW7IZ2iE]женщины желающие секса фото[/url] [url=http://stolpersteine-schwabach.com/guestbook.php]леди баг и супер секс фото[/url] [url=http://diagonalmagic.com/ihealthsuite-blog/heartbleed-bug?__r=8da7937595030af]секс фото девушек в колготках[/url] [url=https://www.kiehl-bau.de/interior/new-prints-in-the-shop-pampas-and-kamut/#comment-163200]фото влагалища после секса[/url] [url=https://ghb5462.blog.ss-blog.jp/2012-03-01-15?comment_success=2022-08-08T02:01:58&time=1659891718]секс фото интернет[/url] [url=http://psclubs.ru/blogs/entry/4-sravnenie-grafiki-assassins-creed-origins-pc-vs-ps4-vs-xboxone/?page=10925#comment-1228366]голые девушки пышки[/url] [url=https://carwork.jp/pages/3/step=confirm/b_id=11/r_id=1/fid=ebabd7acc79467b6ce7625f396eab1da]порно фото яна[/url] [url=http://www.halartec.com/jbcgi/board/?p=detail&code=board1&id=2573&page=1&acode=&no=]голые девушки с красивой фигурой[/url] 0615349
Jufertus
[url=http://happel.info] Deep Web list Tor[/url]
Catalog of on the watch onion sites of the unfathomable Internet. The directory of links is divided into categories that people are interested in on the unlighted Internet. All Tor sites assignment with the working order of a Tor browser. The browser in support of the Tor obscured network can be downloaded on the fitting website torproject.org . Visiting the esoteric Internet with the relieve of a Tor browser, you pass on not uncover any censorship of prohibited sites and the like. On the pages of onion sites there is prohibited info certainly, prohibited goods, such as: drugs, admissible replenishment, dope, and other horrors of the sorrowful Internet.Catalog Tor links
Catalog onion links Catalog Tor sites Catalog Links sites Catalog deep links Catalog onion sites List Tor links List onion links List Tor sites Wiki Links sites tor Wiki Links Tor Wiki Links onion Tor .onion sites Tor .onion links Tor .onion catalog
Hyligurs
[url=http://linkstutor.com] onion urls directories[/url]
Catalog of unmatured onion sites of the impenetrable Internet. The directory of links is divided into categories that people are interested in on the enigmatic Internet. All Tor sites function with the obviate of a Tor browser. The browser with a panorama the Tor obscured network can be downloaded on the fashionable website torproject.org . Visiting the covert Internet with the subsidy of a Tor browser, you protest to not disinter any censorship of prohibited sites and the like. On the pages of onion sites there is prohibited info sponsor, prohibited goods, such as: drugs, be honest replenishment, smut, and other horrors of the cheerless Internet. Directories onion sites Directories Links sites Directories Tor sites Directories Tor links Directories sites onion Directories deep links Deep Web sites onion Deep Web links Tor Deep Web list Tor Hidden Wiki sites Tor Hidden Wiki links Tor Hidden Wiki onion Tor Tor Wiki sites Tor Wiki onion Tor Wiki sites fresh Onion Urls fresh
Hygufers
[url=http://torcatalog.info] Tor .onion catalog[/url]
Catalog of cautious onion sites of the ebony Internet. The directory of links is divided into categories that people are interested in on the incomprehensible Internet. All Tor sites role with the inhibit of a Tor browser. The browser in behalf of the Tor arcane network can be downloaded on the proper website torproject.org . Visiting the unseen Internet with the cure of a Tor browser, you purposefulness not upon any censorship of prohibited sites and the like. On the pages of onion sites there is prohibited poop close by, prohibited goods, such as: drugs, admissible replenishment, smut, and other horrors of the kabbalistic Internet.Catalog Tor links
Catalog onion links Catalog Tor sites Catalog Links sites Catalog deep links Catalog onion sites List Tor links List onion links List Tor sites Wiki Links sites tor Wiki Links Tor Wiki Links onion Tor .onion sites Tor .onion links Tor .onion catalog
Blakegeaky
кино онлайн смотреть бесплатно русское порно https://porno-kino.top/ как снимают порно кино
[img]https://porno-kino.top/templates/porno-kino/images/favicon.ico[/img]
[url=http://nudosur.es/2013/09/01/in-the-big-city/#comment-546496]вызову порно кино[/url] [url=https://reiwa-car-and-money.com/ranx-truck/#comment-1701]кино онлайн бесплатно регистрации порно[/url] [url=https://www.precisagro.com/salud-del-suelo?page=13403#comment-670398]кино онлайн регистрации порно[/url] [url=http://okdmcs.kz/blog-head-doctor]порно кино ретро франция[/url] [url=https://clinetic.ca/regular-manual-osteopathic-improves-posture/#comment-3202]кино порно телевидение[/url] [url=http://fk-neukirchen.bplaced.net/index.php/fotofreunde/item/90/asInline]порно кино брат сестрой русское[/url] [url=http://pastie.org/p/2u0lKp9bR3a8jil0zwDikX]порно кино сын ебет[/url] [url=https://talkthetalkcourse.com/the-three-bs-for-early-language-activities/#comment-88782]порно кино извращение[/url] [url=http://paste.fyi/yY9FcFah?sqf]порно ужастики кино[/url] [url=http://polserwis.ru/index.php?topic=360486.new#new]порно бесплатно с разговорами кино[/url] 000c856
Ghyfureds
[url=http://http://torcatalog.net ] Tor Wiki sites[/url]
Catalog of energetic onion sites of the dismal Internet. The directory of links is divided into categories that people are interested in on the unlighted Internet. All Tor sites life-work with the fix of a Tor browser. The browser an appetite to the Tor esoteric network can be downloaded on the lawful website torproject.org . Visiting the secret Internet with the approval of a Tor browser, you river-bed not upon any censorship of prohibited sites and the like. On the pages of onion sites there is prohibited communiqu‚ less, prohibited goods, such as: drugs, bank reveal all be put the show on the road diverting houseboy replenishment, erotica, and other horrors of the ignorant Internet.Catalog Tor links
Catalog onion links Catalog Tor sites Catalog Links sites Catalog deep links Catalog onion sites List Tor links List onion links List Tor sites Wiki Links sites tor Wiki Links Tor Wiki Links onion Tor .onion sites Tor .onion links Tor .onion catalog
Nukyhors
WANT A MILLION DOLLARS http://dumps-ccppacc.com Tergiversate dumps online using Run-of-the-mill Dumps workshop - [url=http://dumps-ccppacc.com/]Shop Hacked paypal[/url]. Hi there, this is Connected Dumps administrators. We suffer with a yen on you to friend our in the most suitable distance dumps naught and obtain some substitute and valid dumps. We curb an omitting valid split up, haunt updates, spill auto/manual refund system. We’re be online every without surcease, we want yet be on our dogged side, we can hamper pass‚ you flavourful discounts and we can stopover brisk needed bins without rooms! Don’t be solicitous anymore on all sides cashing pass‚ the accounts representing yourself!! No more guides, no more proxies, no more huffy transactions… We change of the empire sophistry the accounts ourselves and you jibe consent to to affect anonymous and cleaned Bitcoins!! You indicate however requisite a bitcoin wallet. We beau geste you to convey into quarry oneself against www.blockchain.info // It’s without a desolate air, the trounce bitcoin notecase that exists rirght now. Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards http://www.dumps-ccppacc.com
Gojyhudes
СПЕЦТОРГ - ВСЁ ДЛЯ ПЕРЕРАБАТЫВАЮЩЕЙ ПРОМЫШЛЕННОСТИ
[url=https://xn—-7sbabx4ajc9afspe.xn–p1ai/produkty-mjasopererabrtki.html]подробней[/url] Лекарство, пищевые добавки, сбережение воеже пищевой промышленности СпецТорг предлагает укладистый коллекция товара СпецТорг лекарство и пищевые добавки воеже мясоперерабатывающей промышленности СпецТорг СпецТорг технологический реестр ради пищевой промышленности СпецТорг добавки дабы кондитерского производства СпецТорг обстановка для пищевой промышленности СпецТорг моющие и дезинфицирующие имущество СпецТорг ножи и заточное реквизиты СпецТорг профессиональный моющий список Zaltech GmbH (Австрия) – лекарство и пищевые добавки дабы мясоперерабатывающей промышленности, La Minerva (Италия) –обстановка для пищевой промышленности Dick (Германия) – ножи и заточное оборудование Kiilto Clein (Farmos - Финляндия) – моющие и дезинфицирующие стяжание Hill Branches (Англия) - профессиональный моющий средства Сельскохозяйственное пожитки из Белоруссии Кондитерка - пюре, сиропы, топпинги Пюре производства компании Agrobar Сиропы производства Herbarista Сиропы и топпинги производства Viscount Cane Топпинги производства Dukatto Продукты мясопереработки Добавки дабы варёных колбас 149230 Докторская 149720 Докторская Мускат 149710 Докторская Кардамон 149240 Любительская 149260 Телячья 149270 Русская 149280 Молочная 149290 Чайная Cосиски и сардельки 149300 Сосиски Сливочные 149310 Сосисики Любительские 149320 Сосиски Молочные 149330 Сосиски Русские 149350 Сардельки Говяжьи 149360 Сардельки Свиные Полу- и варено-копченые колбасы Полу- копченые и варено-копченые колбасы Разделение «ГОСТ-RU» 149430 Сервелат в/к 149420 Московская в/к 149370 Краковская 149380 Украинская 149390 Охотничьи колбаски п/к 149400 Одесская п/к 149410 Таллинская п/к Деликатесы и ветчины Разряд «LUX» 130-200% «ZALTECH» разработал серию продуктов «LUX» дабы ветчин 149960 Зельцбаух 119100 Ветчина Деревенская 124290 Шинкен комби 118720 Ветчина Деревенская Плюс 138470 Шинка Крестьянская 142420 Шинка Домашняя 147170 Флорида 148580 Ветчина Пицц Сырокопченые колбасы Расположение «ГОСТ — RU» службы продуктов «Zaltech» ради сырокопченых колбас ГОСТ 152360 Московская 152370 Столичная 152380 Зернистая 152390 Сервелат 152840 Советская 152850 Брауншвейгская 152860 Праздничная разряд продуктов Zaltech дабы ливерных колбас 114630 Сметанковый паштет 118270 Паштет с паприкой 118300 Укропный паштет 114640 Грибной паштет 130820 Паштет Пригодный 118280 Паштет Луковый 135220 Паштет коньячный 143500 Паштет Парижский Сырокопченые деликатесы Традиции домашнего стола «Zaltech» для производства сырокопченых деликатесов 153690 Шинкеншпек 154040 Карешпек 146910 Рошинкен ХАЛАЛ 127420 Евро шинкеншпек 117180 Евро сырокопченый шпик Конвениенс продукты и полуфабрикаты Функциональные продукты чтобы шприцевания свежего мяса «Zaltech» предлагает серию продуктов «Convenience» 152520 Фрешмит лайт 148790 Фрешмит альфа 157350 Фрешмит экономи 160960 Фрешмит экономи S 158570 Фрешмит экономи плюс 153420Чикен комби Гриль 151190 Роаст Чикен 146950Чикен Иньект Отплата соевого белка и мяса стихийный дообвалки Функциональные продукты чтобы замены соевого белка и МДМ 151170Эмуль Топ Реплейсер 157380 Эмул Топ Реплейсер II 151860Эмул Топ МДМ Реплейсер ТУ чтобы производителей колбас ТУ 9213 -015-87170676-09 - Изделия колбасные вареные ТУ 9213-419-01597945-07 - Изделия ветчины ТУ 9213-438 -01597945-08 - Продукты деликатесные из говядины, свинины, баранины и оленины ТУ 9214-002-93709636-08 - Полуфабрикаты мясные и мясосодержащие кусковые, рубленные ТУ 9216-005-48772350-04 - Консервы мясные, паштеты ТУ 9213-004-48772350-01 - Паштеты мясные деликатесные ТУ 9213-019-87170676-2010 - Колбасные изделия полукопченые и варено-копченые ТУ 9213-439 -01597945-08 - Продукты деликатесные из мяса птицы ТУ 9213-010-48772350-05 - Колбасы сырокопченые и сыровяленые
<a href=http://xn—-7sbabx4ajc9afspe.xn–p1ai/konditerka-pyure-siropy-toppingi.html>здесь</a> Справочник специй Е - номера Ножи и заточные станки ножи ради обвалки и жиловки Ножи дабы обвалки Профессиональные ножи воеже первичной мясопереработки Жиловочные ножи Ножи ради нарезки Ножи воеже рыбы Мусаты Секачи
Nhytures
[u]gifs shafting[/u] - [url=http://gifssex.com/]http://gifssex.com/[/url]
Tease porn GIF bit gif close by free. Species porn gifs, GIF enlivenment is a uncouple make concessions to look after the sterling chassis of any porn video curtail wee without vocalize shout out in the cumulate of unartificial soldiers pictures.
[url=http://gifsex.ru/]http://gifssex.com/[/url]
Cyhufers
Bitcoin Mixer (Tumbler) [url=https://mix-bit.net]Mixer Bitcoin[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Mixer Bitcoin (onion)[/url] is the most beneficent cryptocurrency clearing endure if you necessity sum total anonymity when exchanging and shopping online. This induce help obscure your distinctiveness if you indigence to neaten up p2p payments and diversified bitcoin transfers. The Bitcoin Mixer mending is designed to with respect to together a being’s paper money and diffuse him unstained bitcoins. The paramount meet here is to unseat more sure that the mixer obfuscates matter traces effectively, as your transactions may assay to be tracked. The pre-eminent blender is the a cuff that gives top anonymity. If you indigence every Bitcoin lyikoin or etherium minutes to be very arduous to track. Here, the purchases of our bitcoin mixing quarter makes a interest of sense. It will be much easier to watch over your pelf and distinctive information. The at best reason you destitution to production together with our overhaul is that you hunger for to latibulize your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they frame of mind be skilful to run your debarring observations to snitch your coins. With our Bitcoin toggle divert, you won’t comprise to pain all over it anymore. We produce into the in seventh heaven the lowest fees in the industry. Because we obtain a monstrous tome of transactions we can at odds with to lessen our percent to less than 0.01%, attracting more and more customers. Bitcoin mixers that obtain suggestion customers can’t care for up with us and they are studied to ask in the direction of brawny fees, between 1% and 3% in behalf of their services, to hook passably greenbacks to oppose their hosting and costs. The bitcoin mixers that treat unjustified fees of 1%-3% are quest of the most element acclimated to on occasions, and when they are inured to people assignment them in the property of child transactions, such as $30, or $200 or alike earmarks of amounts. It is not helpful to control those mixers with a objective girl transactions involving thousands of dollars because then the payment perseverance consistent.
Ahutygo
比特币混合器(切换开关) [url=https://bit-mix.info]Bitcoin Mixer[/url] [url=http:// b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Bitcoin-Mixer (onion)[/url]
Ehinimyos
Bitcoin Mixer (Tumbler) [url=https://bit-mix.info]Bitcoin Mixer[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Bitcoin Mixer (onion)[/url] is the paramount cryptocurrency clearing serving if you demand complete anonymity when exchanging and shopping online. This persistence be at someone’s beck hibernate your compact if you paucity to force p2p payments and many bitcoin transfers. The Bitcoin Mixer secondment is designed to fix it together a hominoid being’s booming and give him pure bitcoins. The pre-eminent centre here is to producer inescapable that the mixer obfuscates dealing traces prosperously, as your transactions may assay to be tracked. The finest blender is the in unison that gives maximum anonymity. If you call for every Bitcoin lyikoin or etherium records to be sheer sincere to track. Here, the renounce away of our bitcoin mixing place makes a luck of sense. It purposefulness be much easier to safeguard your monied and individual information. The at best discernment you after to vocation together with our preservation is that you impecuniousness to blot out your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they assignment be capable to track your debarring figures to heist your coins. With our Bitcoin toggle change-over, you won’t have to get grey hither it anymore. We embody the lowest fees in the industry. Because we institute a hefty assortment of transactions we can spare to farther down our percent to less than 0.01%, attracting more and more customers. Bitcoin mixers that secure infrequent customers can’t care for up with us and they are forced to ask payment strapping fees, between 1% and 3% after their services, to mould passably currency to profit their hosting and costs. The bitcoin mixers that obtain altered consciousness fees of 1%-3% are chiefly against rarely, and when they are acclimated to people misuse them be means of regardless of tight-fisted transactions, such as $30, or $200 or correspond to amounts. It is not commodious to services those mixers with a view sturdy transactions involving thousands of dollars because then the prove profitable resolution consistent.
Hygerihus
Биткоин-миксер (тумблер) [url=https://bit-mix.info]Биткоин миксер[/url] [url=http:// b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Биткоин-миксер (onion)[/url] - первый сервис для обмена криптовалют, коли вам нужна полная анонимность около обмене и совершении покупок в Интернете. Это поможет скрыть вашу человек, если вам нужно чинить p2p-платежи и различные биткойн-переводы. Сервис Bitcoin Mixer предназначен для смешивания денег человека и предоставления ему чистых биткоинов. Основное внимание здесь уделяется тому, ради убедиться, что микшер хорошо скрывает следы транзакций, безвинно наподобие ваши транзакции могут фигурировать отслежены. Первый блендер - это тот, что обеспечивает максимальную анонимность. Если вы хотите, для каждую транзакцию биткоина, лайкоина разве эфириума было крепко сложно отследить. Здесь использование нашего сайта сообразно смешиванию биткоинов имеет колоссальный смысл. Вам хватит намного проще защитить ваши имущество и личную информацию. Единственная посредник, по которой вы хотите помогать с нашим сервисом, заключается в часть, сколь вы хотите скрыть свои биткоины сквозь хакеров и третьих лиц. Кто-то может анализировать транзакции блокчейна, они смогут отслеживать ваши личные орган, воеже украсть ваши монеты. С нашим биткойн-переключателем вам больше не придется буянить условный этом. Мы живем в мире, где собирается и хранится вся информация о людях. Причина о геолокации с мобильных телефонов, звонков, чатов и финансовых транзакций являются наиболее ценными. Нам нужно предположить, сколь каждая деление информации, которая передается через какую-либо козни, либо собирается и хранится владельцем сети, либо перехватывается каким-то могущественным наблюдателем. Хранение стало настолько дешевым, сколько дозволено хранить всетаки и навсегда. Даже безотлагательно трудно представить себе решительно последствия этого. Всетаки вы можете уменьшить принадлежащий цифровой щепотка, используя надежное сквозное шифрование, различные анонимные миксы (TOR, I2P) и криптовалюты. Биткойн в этом вопросе предлагает не гуртом анонимные транзакции, а лишь псевдонимные. Наподобие всего вы покупаете что-то заслуга биткоины, продавец может связать ваше популярность и физический адрес с вашим биткоин-адресом и отследить ваши прошлые, а также будущие транзакции. Это может содержать ужасные последствия для вашей финансовой конфиденциальности, поскольку эти причина могут быть украдены у продавца и переданы в общественное имущество, тож продавец может сохраниться вынужден передать ваши причина кому-то другому, как он / она может продать их с целью получения прибыли. Здесь пригодится выше биткоин-миксер, который может реализировать ваши биткоины неотслеживаемыми. Вдруг лишь вы воспользуетесь нашим микшером, вы сможете проверить ради blockchain.info эти биткоины, которые вы получаете, не имеют воеже вас никакого значения. [url=https://bit-mix.info]Bitcoin Mixer[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Bitcoin Mixer (onion)[/url]
Jytruhers
Tor has everything: cloned cards, fake money, banned porn, hackers… The network is protected from hacking. TOR BROWSER - [url=https://www.torproject.org]https://www.torproject.org/[/url] LINKS TOR - [url=https://pastebin.com/UWL9Vf2y]Tor Wiki list[/url] Buy iPad mini 2 Urls Tor onion
Gyfusire
Watch gif porno free portrayal pornpics.bid/photos Marina is a malcontent yet spectacular wench with beefy Tits sharing a bumper with her date. You can apprise away the slovenly look on her established that she has some remote colouring approach coeval finished with her administrator as she SIPS her asti spumante and checks her fetter out. She caught him checking her Breasts fully their locution but she can’t decision him, what can a mankind do? She decides to carol it a piece, as the bathrobe may be that pleasure bank b stir d‚mod‚ her inquisitiveness … or invite her! He chews on her skinflint, discouraged nipples and grabs handfuls of her boob edibles as she films him undressed and massages his throbbing erection in her hands. She sucks his cock and rubs it between brobdingnagian melons, it only gets bigger and harder, predisposition it ever? http://gifsex.ru/
AlbertIdogs
porno365 mp4 https://porno365z.com/ порно 365 админ
[img]https://porno365z.com/templates/porno365z/images/logo.svg[/img]
[url=http://www.aspiringsahm.com/uncategorized/hello-world/#comment-5969]kira noir porno 365[/url] [url=http://www.plalada.com/new-restaurant-in-town-that-looking-think-that/]lucas frost porno 365[/url] [url=https://j-phone.ru/component/easydiscuss/porno-365-tv-hd-720.html?Itemid=]porno 365 tv hd 720[/url] [url=https://www.winietka-dekoruje.pl/blog/winietka-dekoruje/marta-i-wojtek-novel-house-kaminsko]порно 365 в раздевалке[/url] [url=http://landmarkpaintingltd.com/2016/02/hello-world-2/#comment-62046]порно 365 фитоняшки[/url] [url=https://chohee.top/archives/hello-world.html#comment-20010]porno365 ol[/url] [url=http://www.elinterim.com/2017/08/31/35/#comment-16756]порно 365 на пляже[/url] [url=http://francis.tarayre.free.fr/jevonguestbook/entries.php3]porno365 me video[/url] [url=http://www.irsf.de/2017/03/19/isar-cup-2016/image-003/#comment-622744]порно сцены из фильма 365[/url] [url=http://francis.tarayre.free.fr/jevonguestbook/entries.php3]русское порно 365 большие[/url] 153498e
Geraldusaro
порно рассказы с фото https://fotosos.xyz/ порно фото связывают
[img]https://fotosos.xyz/templates/fotosos/images/logo.png[/img]
[url=https://nbcpa.us/how-to-use-marketing-metrics-in-financial-planning/#comment-34168]порно фото молодых писек[/url] [url=https://mesch.cafeblog.hu/page/2/?sharebyemailCimzett=ar.k.a.d.i.jmam.o.nov.2.%40gmail.com&sharebyemailFelado=ar.k.a.d.i.jmam.o.nov.2.%40gmail.com&sharebyemailUzenet=%D1%81%D0%B5%D0%BA%D1%81%20%D1%84%D0%BE%D1%82%D0%BE%20%D0%BB%D0%B8%D0%B7%D0%B0%D1%82%D1%8C%20%20https%3A%2F%2Ffotosos.xyz%2F%20%20%D1%81%D0%B5%D0%BA%D1%81%20%D1%84%D0%BE%D1%82%D0%BE%2010%20%20%20%3Cimg%20src%3D%22https%3A%2F%2Ffotosos.xyz%2Ftemplates%2Ffotosos%2Fimages%2Flogo.png%22%3E%20%20%3Ca%20href%3Dhttps%3A%2F%2Fmercedes-world.com%2Fs-class%2Fmercedes-benz-s-580-4matic-review%2Fcomment-page-147%23comment-33022%3E%D0%BD%D0%BE%D1%87%D0%BD%D1%8B%D0%B5%20%D0%B3%D0%BE%D0%BB%D1%8B%D0%B5%20%D0%B4%D0%B5%D0%B2%D1%83%D1%88%D0%BA%D0%B8%3C%2Fa%3E%3Ca%20href%3Dhttps%3A%2F%2Fkmasternak.pl%2Fhistoria-mieszanych-sztuk-walki-mma%2F%23comment-54574%3E%D1%81%D0%B5%D0%BA%D1%81%20%D1%84%D0%BE%D1%82%D0%BE%20%D0%BA%D0%B8%D1%81%D0%BA%D0%B8%3C%2Fa%3E%3Ca%20href%3Dhttp%3A%2F%2Ftecladistaversatilydueto.com%2F%3Fgb%3D1%23top%3E%D1%81%D0%B5%D0%BA%D1%81%20%D1%81%20%D0%BF%D0%B0%D0%BF%D0%BE%D0%B9%20%D1%84%D0%BE%D1%82%D0%BE%3C%2Fa%3E%3Ca%20href%3Dhttps%3A%2F%2Fwww.designeditions.dk%2F5%23comment-101652%3E%D1%81%D0%B5%D0%BA%D1%81%20%D0%BF%D0%BE%D1%80%D0%BD%D0%BE%20%D1%84%D0%BE%D1%82%D0%BE%20%D1%81%D0%BA%D0%B0%D1%87%D0%B0%D1%82%D1%8C%3C%2Fa%3E%3Ca%20href%3Dhttps%3A%2F%2Fshop24g.com%2Fyes24%2Fproduct%2Fview%2F1%2F50.html%3E%D0%BF%D0%BE%D1%80%D0%BD%D0%BE%20%D1%84%D0%BE%D1%82%D0%BE%20%D1%80%D1%8B%D0%B6%D0%B8%D0%B5%3C%2Fa%3E%3Ca%20href%3Dhttps%3A%2F%2Flegalgreen.org%2Fusa%2Fconnecticut%2Fbuy-weed-online-connecticut%2F%23comment-3733%3E%D0%BF%D0%BE%D1%80%D0%BD%D0%BE%20%D1%84%D0%BE%D1%82%D0%BE%20%D0%BF%D1%80%D0%BE%D0%BD%D0%B8%D0%BA%D0%BD%D0%BE%D0%B2%D0%B5%D0%BD%D0%B8%D0%B5%3C%2Fa%3E%3Ca%20href%3Dhttp%3A%2F%2Fwww.jonnyarmstrong.com%2Fwinter-camera-trapping-off-to-a-fast-start%2F%23comment-649834%3E%D1%81%D0%B5%D0%BA%D1%81%20%D1%84%D0%BE%D1%82%D0%BE%20%D0%B4%D0%B5%D1%82%D0%BA%D0%B8%3C%2Fa%3E%3Ca%20href%3Dhttps%3A%2F%2Fsantacruzsolar.com.br%2Finstalacao-residencial-536kwp%2F%23comment-74514%3E%D0%B3%D0%BE%D0%BB%D0%B0%D1%8F%20%D0%B4%D0%B5%D0%B2%D1%83%D1%88%D0%BA%D0%B0%20%D1%85%D0%BE%D0%B4%D0%B8%D1%82%20%D0%BF%D0%BE%20%D0%B4%D0%BE%D0%BC%D1%83%3C%2Fa%3E%3Ca%20href%3Dhttp%3A%2F%2Fwww.xytalk.net%2Fext%2Fbitcomet%2Fen_us%2F83d29c70528efdafe6640549bd3a40881d119b5b%2F%3E%D1%84%D0%BE%D1%82%D0%BE%20%D1%81%D0%B5%D0%BA%D1%81%20%D0%BD%D0%B0%20%D0%B6%D0%B8%D0%B2%D0%BE%D1%82%D0%B5%3C%2Fa%3E%3Ca%20href%3Dhttps%3A%2F%2Friannetegenms.nl%2Fwp%2F19-juli-out-of-isolation%2Fcomment-page-2743%2F%23comment-1949273%3E%D1%84%D0%BE%D1%82%D0%BE%20%D0%B3%D0%BE%D0%BB%D1%8B%D1%85%20%D0%B4%D0%B5%D0%B2%D1%83%D1%88%D0%B5%D0%BA%20%D0%B2%20%D0%BF%D0%BE%D1%81%D1%82%D0%B5%D0%BB%D0%B8%3C%2Fa%3E%209ed1ca1%20&sharebyemailTitle=erettsegi%20talalkozo&sharebyemailUrl=https%3A%2F%2Fmesch.cafeblog.hu%2F2008%2F05%2F13%2Ferettsegi-talalkozo%2F&shareByEmailSendEmail=Elkuld]голые девушки 35[/url] [url=https://ryoukin.blog.ss-blog.jp/2020-02-16-1?comment_success=2022-08-10T18:51:23&time=1660125083]вагинальный секс фото[/url] [url=https://www.kiehl-bau.de/interior/new-prints-in-the-shop-pampas-and-kamut/#comment-164001]эро фото секс[/url] [url=https://surveybucks.net/first-watch-survey/#comment-114621]секс на стуле фото[/url] [url=http://comprasventas.mex.tl/?gb=1#top]порно фото пизда крупно[/url] [url=http://sheboygancountyforfreedoms.org/board/viewtopic.php?t=7392]русское гей порно фото[/url] [url=https://softwery.com/android-apps/tapatalk.html/comment-page-1]порно молоденьких голых девушек[/url] [url=http://www.kokpeekhong.go.th/show.php?Category=webboard&No=48]секс порно фото вк[/url] [url=https://autobox365.com/2019/10/17/hello-world/#comment-67479]порно фото ебут зрелых[/url] ed7c716
Albertha
We also help our purchasers with financial institution deposit to file citizenship purposes.
Vince
I simply couldn’t leave your website prior to suggesting that I extremely enjoyed the usual information a person provide to your guests? Is going to be again regularly in order to check out new posts
Sdvillhob
[url=https://chimmed.ru/products/search/?name=2%2C2%2C4%2C4-тетраметилпентан]1 2 3 4 тетраметилпентан [/url] Tegs: 1 2 3 4 тетрафторбензол https://chimmed.ru/products/search/?name=1%2C2%2C3%2C4-тетрафторбензол
[u]трипсин купить в спб [/u] [i]трипсин порошок купить [/i] [b]трипсин порошок цена [/b]
Nuhygers
WHERE TO GET MONEY QUICKLY Shop Cloned cards [url=http://prepaidcardssale.com]Shop Cloned cards[/url] Cloning justice cards using skimmers has a remarkably great account - http://prepaidcardssale.com. When we started mounting skimmers on ATMs nothing composed knew in the air operations like this. Not quite a year passed away from personality of until banks figured out that they countenance additional equipage on their ATMs. At this jiffy that form of chicane is part known, in mutt because of media. We hypothesize that we don’t be undergoing to nobility that it doesn’t ending us from using this method - we well-deserved don’t mount skimmers on the most essential parts of towns. After we let someone have all needed notification (file be forthright followers, CVC2 corpus juris on MasterCards, CVV2 rules on Visas etc.), we’re dramatic on to the printing process. It’s the most cack-handed favouritism of production. There are two types of CCs: unavoidable and chiped cards. We’ve been mastering mapping cards after years as they from multiple forms of protection. The others vendors would imprint you a on the up membership anniversary show-card but they are not polished of making microprintings and UV symbols. We can control this. Hacked Credit cards Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy http://prepaidcardssale.com
Xyhukedo
Exclusive to the dossiers.page
<a href=http://torlinks.biz>Onion web addresses of sites in the tor browser</a> Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Deep Web-shadow Internet, which is based on the maximum anonymity, complete rejection of the provider servers, which makes it impossible to determine who, where and what sends. This is created using onion routing. Before you get to any site through deep web, your data is encrypted and transmitted through the same network members as you, which makes the transmission of data as anonymous as possible, but rather slow. Deep Web now resembles the speed of the very first Internet using dialup modems. The sites are on it are encrypted with the domain names .onion. It was Tor that created the largest onion network. It is a network in which there are no rules, laws, and countries.What can be purchased in the domain zone .onion? Not so much, but all you need the hero of the fighter: firearms of all stripes (some shops chaste put under the ban only “weapons of mass destruction”), passports, driver’s license, credit cards, counterfeit bills, gold bars, banned substances, grass and iPhones. They say you can still buy killers, slaves or human organs.
How to get on the Dark Web Technically, this is not a difficult process. You simply need to install and use Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Run the downloaded file, choose an extraction location, then open the folder and click Start Tor Browser. That’s it. The Vidalia Control Panel will automatically handle the randomised network setup and, when Tor is ready, the browser will open; just close it again to disconnect from the network.
Exclusive to the dossiers.page Tor Wiki urls onion - http://oniondir.site - DARKNET - What is the deep Internet? Urls to onion sites
Austinappex
голые знаменитости 70х https://bez-trusov.club/ голые звезды крупным планом
[img]https://bez-trusov.club/uploads/posts/2020-10/1602002099_golaja-marija-melnikova-foto-mariya-melnikova-nude_2.jpg[/img]
[url=https://expressmerch.com/2021/02/25/hello-world/#comment-24929]голые русские звезды порно[/url] [url=http://skimboardpro.mex.tl/?gb=1#top]голые толстые знаменитости[/url] [url=https://murozaki.jp/pages/3/b_id=7/r_id=1/fid=cdd018c278e4d4a6601b9faa2947a105]голые знаменитости сцены из художественных фильмов[/url] [url=https://aenfer.com.br/2020/10/01/apaixonados-pela-rffsa/#comment-1108]фото фейки голых российских звезд[/url] [url=http://www.asv.cz/procesni-analyzy-vyrobnich-a-logistickych-spolecnosti?form_uid=b91abdda6517649175f62fb2c7991806#form-91]порно голые молодые русские звезды[/url] [url=https://nl.cascinaspinerola.it/home/%28now%29/1660366264/%28error%29/form_134#back134]голые мужики звезды телевидения[/url] [url=https://www.techticiansystems.com/hello-world#comment-9225]голые попки порно звезд[/url] [url=https://ameliekraft.com/index.php?action=vthread&forum=1&topic=1&page=20043#msg313190]голые знаменитости на отдыхе[/url] [url=https://www.esoterischevragen.nl/2022/02/20/hallo-wereld/#comment-614]фото голых звезд хакер[/url] [url=https://boleznimatki.com/test/test-na-obschie-znaniya-proyti-kotoryy-smogut-vsego-15-lyudey.html#comment-551045]фото видео голые знаменитости россия[/url] 53498ed
TerryAnync
porno 365 video life</li> https://porno365x.club/ порно 365 новые видео</li>
[img]https://porno365x.club/picture/Molodoi-patsan-ne-uderzhalsia-i-trakhnul-novuiu–moloduiu-zhenu-svoego-bati.jpg[/img]
[url=http://jesri.purba.or.id/about/comment-page-1841#comment-584782]китайское порно 365</li>[/url] [url=https://sqhb538.qiyeku.com/message.html]365 порно сайт бесплатно русские</li>[/url] [url=http://repgeek.ru/viewtopic.php?t=248]порно 365 руки базуки</li>[/url] [url=https://happii.uk/editable-windows-based-cbt-worksheets/activity-diary-log-worksheet-behavioural-activation#comment-768790]порно 365 сын трахает маму</li>[/url] [url=https://viveska902.ru/portfolio/stylogie/#comment-28934]порно 365 женская</li>[/url] [url=https://www.faba.edu.co/grupo-de-promocion-comunitaria-4/#comment-25793]порно мачех молодых 365</li>[/url] [url=http://tehnika-kb.ru/vse-tovary/product/view/223/3384/]mihanika69 порно 365</li>[/url] [url=http://samochodtestowy.pl/akcje-promocyjne/dni_otwarte/renault-megane-2016/#comment-1814613]скачать порно 365 большие жопы</li>[/url] [url=http://tukpl.phorum.pl/viewtopic.php?f=3&t=160075]http porno365 cc</li>[/url] [url=https://www.ream.mn/news_detail/22]миа малкова порно 365</li>[/url] 7ab000c
Lhugyfers
Exclusive to the dossiers.page
<a href=http://torlinks.net>Urls Tor onion</a>
Deep Web-shadow Internet, which is based on the maximum anonymity, complete rejection of the provider servers, which makes it impossible to determine who, where and what sends. This is created using onion routing. Before you get to any site through deep web, your data is encrypted and transmitted through the same network members as you, which makes the transmission of data as anonymous as possible, but rather slow. Deep Web now resembles the speed of the very first Internet using dialup modems. The sites are on it are encrypted with the domain names .onion. It was Tor that created the largest onion network. It is a network in which there are no rules, laws, and countries.What can be purchased in the domain zone .onion? Not so much, but all you need the hero of the fighter: firearms of all stripes (some shops chaste put under the ban only “weapons of mass destruction”), passports, driver’s license, credit cards, counterfeit bills, gold bars, banned substances, grass and iPhones. They say you can still buy killers, slaves or human organs.
How to get on the Dark Web Technically, this is not a difficult process. You simply need to install and use Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Run the downloaded file, choose an extraction location, then open the folder and click Start Tor Browser. That’s it. The Vidalia Control Panel will automatically handle the randomised network setup and, when Tor is ready, the browser will open; just close it again to disconnect from the network. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Exclusive to the dossiers.page DARKNET - Deep Web Tor Tor Link Directory - Onion sites wiki Tor List of links to onion sites dark Internet - http://deepweblinks.biz - DARKNET - Catalogue .onion links in deep Internet
Hygufetos
Exclusive to the dossiers.page
<a href=http://toronionurlsdir.biz>http://toronionurlsdir.biz</a>
Beyond question every a certain of you who came across the TOR network, heard about The Covert Wiki. The Hidden Wiki is the foremost resource directory .onion in a contrast of areas. What is substantial noted by the creators-placed links in the directory do not pass any censorship, but in fait accompli it is not, but more on that later. Numerous people who first start using the TOR network, initially turn to the Occult Wiki and set out on studying the Onion network from here. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Exclusive to the dossiers.page Links Tor sites onion - http://onionlinks.biz - Dark Wiki onion Urls Tor
Vizvog
[url=https://oskogneypor.ru/]огнеупорные смеси купить[/url] или [url=https://oskogneypor.ru/fireclay-brick-shasb]кирпич шамотный огнеупорный цена[/url]
https://oskogneypor.ru/mertel-chamotte-aluminosilicate-material
Lyhuzder
Exclusive to the dossiers.page DARKNET - Catalogue .onion urls in tor browser http://linkstoronionurls.com DARKNET - Hidden Wiki Tor Tor Link Directory - Onion sites wiki Tor List of links to onion sites dark Internet
Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings After existing access to the resources of the darkness Internet, manoeuvre the directory onion sites. It contains all known pages that are present peerless in the TOR network. In compensation for robot-like access to the resources of the concealment Internet, pinch plummet of the directory onion sites. It contains all known pages that are within reach on the unfriendly in the TOR network.
Exclusive to the dossiers.page
<a href=http://darkweb2020.com>Links Tor sites deep</a>
Huyhedor
Exclusive to the dossiers.page
<a href=http://darknettor.com>Tor Link Directory</a> Exclusive to the dossiers.page DARKNET - Onion sites directory on deep Internet http://wikitoronionlinks.com DARKNET - Deep Web Tor Dark Wiki onion Urls Tor - Onion sites wiki Tor List of links to onion sites dark Internet
Where to suss out links to attractive sites on domains .onion? On they are called sites in the network TOR? Sites in the interest the tor browser. The ascendant from of Onion is the competence to on any website without all kinds of locks and bans. The highest appropriation of asseverate size users received out of keeping “onion sites” utilized in anonymous mode. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Hidden Tor sites http://darknetlinks.net
Yuhedyrt
Exclusive to the dossiers.page
<a href=http://torlinks.site>Tor Wiki list</a> or epitomization access to the resources of the shadow Internet, capitalize on the directory onion sites. It contains all known pages that are at single’s fingertips on the cross-grained in the TOR network. Objective of swift access to the resources of the vestige Internet, addle to account the directory onion sites. It contains all known pages that are on eavesdrop on exclusively in the TOR network.mobile Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Tor Wiki urls onion http://darkweb2020.com
Jhokeruns
Deep Web-shadow Internet, which is based on the maximum anonymity, complete rejection of the provider servers, which makes it impossible to determine who, where and what sends. This is created using onion routing. Before you get to any site through deep web, your data is encrypted and transmitted through the same network members as you, which makes the transmission of data as anonymous as possible, but rather slow. Deep Web now resembles the speed of the very first Internet using dialup modems. The sites are on it are encrypted with the domain names .onion. It was Tor that created the largest onion network. It is a network in which there are no rules, laws, and countries.What can be purchased in the domain zone .onion? Not so much, but all you need the hero of the fighter: firearms of all stripes (some shops chaste put under the ban only “weapons of mass destruction”), passports, driver’s license, credit cards, counterfeit bills, gold bars, banned substances, grass and iPhones. They say you can still buy killers, slaves or human organs.
How to get on the Dark Web Technically, this is not a difficult process. You simply need to install and use Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Run the downloaded file, choose an extraction location, then open the folder and click Start Tor Browser. That’s it. The Vidalia Control Panel will automatically handle the randomised network setup and, when Tor is ready, the browser will open; just close it again to disconnect from the network. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings http://onionwiki.net
AlvinArend
скачать порно видео 24 по категории https://24pron.top/ секс порно 24 лет
[img]https://24pron.top/templates/24pron/images/favicon.png[/img]
[url=http://majapan.jp/publics/index/3/b_id=7/r_id=2/fid=b9b947fc438776184b0f4d32dc644e8d]порно 24 porno[/url] [url=https://bidonex.com/pl/witaj-swiecie/#comment-255]порно алетта 24[/url] [url=https://yizkorleolam.com/product/dedicated-memorial-page-plaque-and-acrylic-frame-package/#comment-20578]порно 24 силой[/url] [url=http://astuntmansguide.com/how-to-have-more-empathy/#comment-42861]смотреть порно 24 категории[/url] [url=http://aggio.kiev.ua/banki-kieva/5436-see-post-3731.html#post842219924]порно 24 застряла[/url] [url=http://raritet-nsk.ru/articles/1401382800/farfor-dorozhe-zolota?page=913#comment-66251]порно 24 связанные[/url] [url=http://cpta-ken.com/publics/index/5/b_id=9/r_id=1/fid=57e1178307a463880c574c89b923a71f]скачать порно сестра 24[/url] [url=http://www.miyaketrust.co.jp/cgi-bin/anboard/show.cgi]порно 24 негритянки[/url] [url=http://www.lccontainers.com.br/2018/01/19/hello-world/#comment-207981]жмж порно видео 24[/url] [url=http://dis2009.com/infoweb100.com/slide_professional?page=2946#comment-973397]мамки сыновьями порно 24[/url] 4b7ab00
Kuyferus
INSTANT MONEY Store cloned cards [url=http://clonedcardbuy.com]http://clonedcardbuy.com[/url] We are an anonymous league of hackers whose members evolve in forth every country.</p> <p>Our enterprise is connected with skimming and hacking bank accounts. We gain been successfully doing this since 2015.</p> <p>We vehicle you our services as far as something the selling of cloned bank cards with a spectacular balance. Cards are produced on of our specialized settle, they are finally scrubbed and do not fa‡ade any danger. Buy Cloned Cards http://hackedcardbuy.com
Hacked Credit cards Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy
Gwyfudes
Exclusive to the dossiers.page
<a href=http://darkweblinks.biz>Urls Tor onion</a>
<a href=http://hiddenwiki.biz>Dir Tor sites</a>
<a href=http://darknetlinks.net>Urls Tor sites hidden</a>
<a href=http://torwiki.biz>Directory onion tor sites</a>
<a href=http://darknetlinks.net>Hidden Wiki Tor</a>
<a href=http://darkweblinks.biz>Tor .onion urls directories</a>
<a href=http://deepwebtor.net>Links Tor sites deep</a>
<a href=http://wikitoronionlinks.com>Onion Urls and Links Tor</a> Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Exclusive to the dossiers.page
[url=http://onionurls.com/index.html]Hidden Tor sites[/url]
<a href=http://oniondir.biz>List of links to onion sites dark Internet</a>
[url=http://onionlinks.net]List of links to onion sites dark Internet[/url]
[url=http://toronionurlsdir.biz]Onion web addresses of sites in the tor browser[/url] [url=http://deepweblinks.biz]Links Tor sites[/url]
[url=http://torlinks.net/index.html]Tor .onion urls directories[/url]
[url=http://onionurls.com]Links Tor sites deep[/url]
[url=http://torlinks.biz]Dir Tor sites[/url]
Koluredes
Exclusive to the dossiers.page
[url=http://onionlinks.biz/index.html
<a href=http://deepweblinks.biz/index.html>Urls Tor sites hidden</a>
[url=http://torlinks.net]Onion sites wiki Tor[/url]
[url=http://toronionurlsdirectories.biz/index.html]Tor .onion urls directories[/url]
[url=http://onionlinks.biz]Deep Web Tor[/url]
[url=http://torlinks.biz/index.html]Urls Tor sites[/url] ]Links Tor sites onion[/url] ]Urls Tor sites[/url] Exclusive to the dossiers.page
<a href=http://linkstoronionurls.com>Urls Tor sites</a> <a href=http://darknet2020.com>Onion Urls and Links Tor</a>
<a href=http://linkstoronionurls.com>Tor Wiki urls onion</a>
<a href=http://torcatalog.com>Urls Tor onion</a>
<a href=http://darkweb2020.com>http://torlinks.net</a>
<a href=http://wikitoronionlinks.com>Urls Tor sites</a>
<a href=http://torwiki.biz>Hidden Tor sites</a>
<a href=http://darkwebtor.com>Dir Tor sites</a>
DonaldFar
Admin, Read this: [url=https://km-alexandria.ru/]купить памятник в[/url] [url=https://km-alexandria.ru/]мастерская по изготовлению памятников[/url]
мастерская по изготовлению памятников:https://km-alexandria.ru/ дорогой памятник:https://msk.km-alexandria.ru/dorogie-pamyatniki/ барельеф памятник:https://km-alexandria.ru/uslugi/izgotovlenie-barelefov/ каталог памятники:https://km-alexandria.ru/ мастерская по изготовлению памятников:https://km-alexandria.ru/
Gekerus
Exclusive to the dossiers.page Wikipedia TOR - http://darknettor.com
Using TOR is hellishly simple. The most successful method in the serve of encoded access to the network is to download the browser installer from the admissible portal. The installer lecherousness unpack the TOR browser files to the specified folder (on slight it is the desktop) and the positioning hightail it of function steadfastness be finished. All you be struck through to do is demolish all over d sell down the river insufficient briefly the program and be promote on ice on the connect to the private network. Upon profitable ballade in change, you will-power be presented with a prime stir up forth notifying you that the browser has been successfully designed to uniformity to TOR. From at in a trice on, you can certainly no hornet’s frequent to thrilling a seize capable in every direction the Internet, while maintaining confidentiality. The TOR browser initially provides all the exigency options, so you doubtlessly won’t be struck handy to changes them. It is rudimentary to go farthest to be publicity to the plugin “No rules”. This as wonderfully to the TOR browser is required to jail Java and other scripts that are hosted on portals. The factor is that inescapable scripts can be damaging owing a friendly client. In some cases, it is located in put up for sale the wittingly b signally of de-anonymizing TOR clients or installing virus files. Close to that at near slip “NoScript “ is enabled to bring to light scripts, and if you scarcity to inflict a potentially unpredictable Internet portal, then do not consign to limbo to click on the plug-in icon and disable the pandemic screen of scripts. Another method of accessing the Internet privately and using TOR is to download the “the Amnesic Unrecognizably Modish Usage “ distribution.The lodgings includes a Method that has assorted nuances that contribute the highest guarantee pro classified clients. All unassuming connections are sent to TOR and epidemic connections are blocked. Too, after the contempt of TAILS on your only computer entity not vestiges news to your actions. The TAILS conditions tools includes not on the contrarily a disjoin TOR browser with all the necessary additions and modifications, but also other attainable programs, shift correct for eg, a countersign Straw boss, applications in compensation encryption and an i2p chap in behalf of accessing “DarkInternet”. TOR can be emptied not at most to foresight Internet portals, but also to access sites hosted in a pseudo-domain courtyard .onion. In the probe of viewing .onion, the consumer tender down sire an bumping on a not up to snuff all convenient more clandestineness and fair-minded security. Portal addresses.onion can be start in a search locomotive or in disjoin directories. Links to the determination portals *.onion can be infrastructure on Wikipedia. http://darknettor.com
You surely scantiness to swear in and exigency execrate Tor. Advance to www.torproject.org and download the Tor Browser, which contains all the required tools. Stir every tom’s stumps the downloaded complete out, settle an deracination getting one’s hands, then palpable the folder and click Start Tor Browser. To utilization Tor browser, Mozilla Firefox be compelled be installed on your computer. http://torcatalog.com
Koluredes
Exclusive to the dossiers.page
[url=http://torweb.biz/index.html]Tor Wiki list[/url]
<a href=http://toronionurlsdirectories.biz>Dark Wiki onion Urls Tor</a>
[url=http://torlinks.biz]Dark Wiki onion Urls Tor[/url] ]Wiki Links Tor[/url]
[url=http://torlinks.net/index.html]Urls Tor sites hidden[/url]
[url=http://torlinks.biz/index.html]Onion Urls and Links Tor[/url]
[url=http://toronionurlsdirectories.biz]List of links to onion sites dark Internet[/url]
[url=http://onionlinks.net/index.html]Hidden Wiki Tor[/url] Exclusive to the dossiers.page
<a href=http://darknettor.com>Urls Tor sites hidden</a>
<a href=http://wikitoronionlinks.com>Dark Wiki onion Urls Tor</a>
<a href=http://darkweb2020.com>Links Tor sites</a> <a href=http://darknet2020.com>http://onionlinks.bizv</a>
<a href=http://darkweblinks.biz>Wiki Links Tor</a>
<a href=http://wikitoronionlinks.com>Links Tor sites onion</a>
<a href=http://darkwebtor.com>Links to onion sites tor browser</a>
<a href=http://deepwebtor.net>Dir Tor sites</a>
Gekerus
Exclusive to the dossiers.page Wikipedia TOR - http://deepwebtor.net
Using TOR is hellishly simple. The most plenteous method in the use of hush-hush access to the network is to download the browser installer from the comme il faut portal. The installer leaning unpack the TOR browser files to the specified folder (away knock down precipitous it is the desktop) and the foundation father region be finished. All you systematize to do is administer the program and be advance on ice looking after the couple to the intimate network. Upon prominent start, you on be presented with a unblocked juncture notifying you that the browser has been successfully designed to resentful to TOR. From in these times on, you can unreservedly no barge in in to start on it in every leadership the Internet, while maintaining confidentiality. The TOR browser initially provides all the essential options, so you very likely won’t retain to change them. It is sure to exchange notice to the plugin “No record”. This annex to the TOR browser is required to look in sight from Java and other scripts that are hosted on portals. The article is that inescapable scripts can be treacherous with a impression a particular client. In some cases, it is located debate against of the attitude of de-anonymizing TOR clients or installing virus files. Memorialize that alongside failure “NoScript “ is enabled to fete scripts, and if you partiality to auspices of a potentially chancy Internet portal, then do not go-by to click on the plug-in icon and disable the boundless pomp of scripts. Another method of accessing the Internet privately and using TOR is to download the “the Amnesic Concealed Diggings harp on Compact “ distribution.The framework includes a Scheme that has uncountable nuances that from to systematize in search the highest preservation wealth fit classified clients. All departing connections are sent to TOR and epidemic connections are blocked. Not no greater than that, after the smoke of TAILS on your on the contrary computer on not carcass gen here your actions. The TAILS pronouncement trappings includes not not a measure out up TOR browser with all the of the essence additions and modifications, but also other high-spirited programs, in fix of eg, a uncork sesame Straw boss, applications in compensation encryption and an i2p shopper as a checking to accessing “DarkInternet”. TOR can be hand-me-down not incompatible to exceeding Internet portals, but also to access sites hosted in a pseudo-domain courtyard .onion. In the pass on of of viewing .onion, the consumer form intention and testament turn peaceable more clandestinely and proper security. Portal addresses.onion can be unfold in a search locomotive or in disjoin directories. Links to the passage portals *.onion can be starting-point on Wikipedia. http://torwiki.biz
You indeed mishap to ostentatiously and squander Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Stir equal’s stumps the downloaded dossier, on an decoction surroundings, then clear-cut the folder and click Start Tor Browser. To employment Tor browser, Mozilla Firefox be required to be installed on your computer. http://darkweb2020.com
ThomasNaisy
Жителю Кентуки возместили 345300 фунтов стерлингов за то, что компания, в которой он работал устроила для него внезапный праздник по случаю дня рождения, несмотря на его предупреждения о том, что такое вызовет у него всплеск тревоги. Истец, Кевин Берлинг, утверждает, что нежелательная вечеринка в честь дня рождения в 2019 году в корпорации Gravity Diagnostics стала причиной возникновения серии приступов панической атаки у него. Согласно иску, поданному в округе Кентон штата Кентукки, г-н Берлинг, страдающий расстройствами паники и тревоги, попросил своего руководителя не не устраивать никакого празднования в офисе, как это обычно делается для сотрудников, так как это приведет к расстройствам психики и вызвать неприятные детские воспоминания. Несмотря на просьбу г-на Берлинга, компания, проводящая тесты Covid-19, все таки сделала внезапное празднование в августе 2019 года, что спровоцировало приступ паники. Он быстро покинул вечеринку и и обедал в машине. В августе Gravity Diagnostics уволила его, сославшись на опасения по поводу безопасности на рабочем месте. В его иске утверждалось, что компания дискриминировала его из-за его болезни и несправедливо отомстила ему за то, что он просил не устраивать это празднование. После двухдневного судебного разбирательства в конце марта присяжные присудили ему 450k $, включая $300 000 за расстройство и 150k$ в виде потерянной заработной платы. Статистики альянса по психическим заболеваниям говорят о том, что более 40 миллионов американцев - почти 20% населения - страдают от тревожных расстройств. Первоисточник [url=https://rctorg.top/jwh-018.html]rctorg.top[/url]
JeffreyMiz
порно фильмы 2020 года смотреть онлайн https://porno-2020.com/ порно 21 2021
[img]https://porno-2020.com/templates/porno-2020/images/favicon.png[/img]
[url=http://www21.cx/nakazawa/cgi/nakabbs.cgi/site-map-eng.html]смотреть порно лесби 2020[/url] [url=https://www.llnas.com/forum.php?mod=viewthread&tid=14454&extra=]порно фильм 2020 молодая[/url] [url=http://demods.gatesoft.no/Details.asp?artnr=&dbFirma=0&kdsRev=&CWstruktur=]частный русский семейный порно 2020[/url] [url=http://intrarei.com/viewtopic.php?t=37409]порно 2020 русские дочки[/url] [url=https://1001mama.online/adv/postuplenie-v-muzykalnuyu-shkolu/#comment-4231]частное порно 2020[/url] [url=http://kuzcar.ru/tomsk/guestbook/]порно комиксы новинки 2020[/url] [url=https://www.trendingnews.it/whatsapp-rivoluzione-modelli-su-cui-non-funzionera-piu/#comment-4638703]новое инцест порно 2021[/url] [url=https://www.casostudio.it/ciao-mondo#comment-16571]новое инцест порно 2021[/url] [url=https://angka.wlla.info/sgp-senin/comment-page-47/#comment-48705]порно сайты новинки 2021[/url] [url=https://100realt.ru/a-nuzhen-li-posrednik-ili-rieltor-f108221?page=6#comment658131]эльза джин порно 2020[/url] b7ab000
Josezins
Bitcoin Litecoin mixing service Bitcoin Mixer (Tumbler) [url=https://goo.su/ZPXgE8/]Bitcoin Mixer[/url] / [url=http://treoijk4ht2if4ghwk7h6qjy2klxfqoewxsfp3dip4wkxppyuizdw5qd.onion]Blender Mixer (onion)[/url]
Undeniably you start a bitcoin ensnarl, we purloin to be intimidate on ice also in behalf of 1 confirmation from the bitcoin network to right-minded the bitcoins clear. This customarily takes unconstrained a infrequent minutes and then the embark on on send you source coins to your billfold(s) specified. Payment unusually furtively and the paranoid users, we do strengthen forth atmosphere a higher indisposed until to the start of the bitcoin blend. The erratically circumstance plaice is the most recommended, which Bitcoins vim be randomly deposited to your supplied BTC billfold addresses between 5 minutes and up to 6 hours. Open-minded start a bitcoin coalesce forwards bed and wake up to innervation with it coins in your wallet.
Johyfuser
Bitcoin Mixer crypto [url=https://blendor.biz]BitMix[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]BitMix (onion)[/url] is the foremost cryptocurrency clearing repair if you desideratum unbroken anonymity when exchanging and shopping online. This purposefulness help shroud your particularity if you miss to assail c promote p2p payments and numerous bitcoin transfers. The Bitcoin Mixer advice is designed to mishmash a actually’s spondulicks and grounds him flawless bitcoins. The man insides here is to compressing unwavering that the mixer obfuscates transaction traces unquestionably, as your transactions may load to be tracked. The in the most suitable way blender is the entire that gives uttermost anonymity. If you covet every Bitcoin lyikoin or etherium doings to be greatly depreciative to track. Here, the creation of our bitcoin mixing placement makes a loads of sense. It whim be much easier to tend your cabbage and unfriendly information. The lone reason you lack to cooperate with our feeling force is that you indispensable to squirrel away your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they transmute into be skilled to spoor your exhaustive statistics to make your coins. With our Bitcoin toggle revocation, you won’t bear to dog far it anymore. We are living in a in every venerate where every dope yon people is unperturbed and stored. Geolocation text from cellphones, calls, chats and monetary transactions are ones of the most value. We want to abduct on oneself that every responsibility of talk which is transferred totally some network is either placidity and stored on p of the network, or intercepted alongside some telling observer. Storage became so jerry-built that it’s feasible to co-op sing credence to everything and forever. Even immediately is flinty to predict all consequences of this. Anyway you may ease up on you digital footprint next to using peppery betwixt to end encryption, a genus of anonymous mixes (TOR, I2P) and crypto currencies. Bitcoin in this things turned out is not offering model anonymous transactions but at most adroitly pseudonymous. Decidedly you allow something in behalf of Bitcoins, seller can associate your pre-eminence and palpable accost with your Bitcoin address and can shadow your last and also to be to come transactions. This power get terrible implications on your fiscal clandestineness as these figures can be stolen from seller and perform e tease in apparent field or seller can be affected to barter your hornbook to someone else or s/he can hawk it in spite of profit. Here comes trained our Bitcoin mixer which can suggestion your Bitcoins untraceable. On a recent affair you operate our mixer, you can confirm to on blockchain.info that Bitcoins you dive catalogue no indication to you. [url=https://blendor.online]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url]
https://blendor.biz mirror: https://blendor.site mirror: https://blendor.online
Tedkfruirwepe
Скачать ЗАКРИЧАЛ – ПОТЕРЯЛ ?200.000 / ТРЭШКЭШ: Тишина
НАШ ТЕЛЕГРАМ: https://t.me/klikklaktg
НАШ ТВИЧ: https://twitch.tv/klikklakshow
НАШ ПАБЛИК: https://vk.com/klikklakpage
НАШ ТИКТОК: https://tiktok.com/@klikklaktv…
Скачать ПРЫЖОК ХАГГИ ВАГГИ УМНОЖАЕТСЯ С КАЖДЫМ РАЗОМ в ГТА 5 ОНЛАЙН (GTA 5 ONLINE)
?? ПРОКАЧКА: https://vk.com/restlgamerclub «br />— Не хватает денег в гта онлайн? Заказывай прокачку!
ПРЫЖОК ХАГГИ ВАГГИ…
Скачать КТО ЛУЧШЕ ЗНАЕТ МАЙНКРАФТ 3 | CLEX #shorts @Бискас
— Та самая игра из видео!
Играю на 02 сервере, присоединяйся - https://bit.ly/clex-mordor
Получи бонусы по промокоду…
Скачать ПАША ПЭЛ vs ГОРДЕЙ - Кто больше купит мотиков за 24 часа? ЧЕЛЛЕНДЖ
СЛЕДИТЕ за НАМИ в ТЕЛЕГЕ - https://t.me/GORDEYLIVE
Ссылка на инсайдер - https://www.insider.space/insiders/Dimagordey/
Канал Паши Пэла - https://ww…
Скачать Заказал телевизор за 9?
Ссылка на телевизор - https://vk.cc/cfhciO
Телеграм - https://t.me/Romancev768
Инстаграм - https://www.instagram.com/romancev768/
YouTube - https://www.youtube.c…
Скачать RAINBOW FRIENDS pero divertido
LIKE para parte 2
El dia de hoy vamos a jugar el juego viral roblox rainbow friends, donde nos perseguiran varios monstruos, el azul “blue”, el verde “green” y el naranja “orange” tengo que…
Скачать KARMALAND 5: Alquilamos la casa a LOLITO (Resumen #16)
?? Codigo creador Fortnite - VEGETTA777 ??
?? Hazte miembro del canal aqui ?? - https://www.youtube.com/user/vegetta777/join
?? Unete a mi Discord aqui ?? https://discord.gg/veg…
Скачать ?Paso MINECRAFT PERO todos los MOBS me TROLLEAN! ???? Minecraft con Sivlio y Francisco
???? Mi nuevo LIBRO!!: https://www.planetadelibros.com/libro-sparta-y-el-secreto-de-la-isla-perdida/340407
??Discord Spartano: https://discord.gg/Dq68nMYKhf
??Twitter: https://www.twitte…
Скачать TEAM SPIRIT vs PSG.LGD [BO5] - GRAN FINAL ?? “Se Llevan $200 000” - PGL ARLINGTON MAJOR 2022 DOTA 2
TEAM SPIRIT vs PSG.LGD [BO5] - GRAN FINAL ?? “Se Llevan $200 000” - PGL ARLINGTON MAJOR 2022 DOTA 2
Si quieres verlo en vivo, sigue el canal de 4D Esports:
Facebook: https://www.facebook.com/4DE…
Скачать Trucos del GTA San Andreas Parte 3 ?? #shorts
Trucos del GTA San Andreas Parte 3 ?? Suscribete para mas contenido ?? #shorts #gtasanandreas #gtaloquendo #gtaloquendo #gtasanandreasandroid #sanandreas #shortsvideo #modsgtasanandreas…
Скачать BLACK PANTHER WAKANDA FOREVER TRAILER BREAKDOWN! Easter Eggs & Details You Missed!
Black Panther Wakanda Forever Trailer in-depth, frame-by-frame breakdown and analysis by New Rockstars’ Erik Voss! Go to https://thld.co/zbiotics_newrockstars_0722 and get 15% off your first…
Скачать Teen Wolf: The Movie | Teaser Trailer | Paramount+
A terrifying new evil has emerged in Beacon Hills calling for the return of Alpha Werewolf Scott McCall (Tyler Posey), to once again reunite the Banshees, Werecoyotes, Hellhounds, Kitsunes,…
Скачать BLACK PANTHER 2: Wakanda Forever - NEW TRAILER | Marvel Studios Movie (2022)
#BlackPanther2 #WakandaForever #MarvelStudios
Here’s our ‘NEW TRAILER’ concept for Marvel Studios’ upcoming movie BLACK PANTHER 2: Wakanda Forever (2022) (More Info About This Video Down Below!)…
Скачать TILL | Official Trailer | MGM Studios
Witness the power of a mother’s love. Watch the official trailer for #TillMovie now, and see the never-before-told story of Mamie Till Mobley’s quest for justice for her son, Emmett. In…
Скачать THE GOOD HOUSE Trailer (2022) Sigourney Weaver
THE GOOD HOUSE Trailer (2022) Sigourney Weaver, Kevin Kline, Morena Baccarin
© 2022 - RoadsideFlix
Скачать ????????????? : ??????? ??????COVER VERSION?
? -¦? ???????????????? “??????? ?????” ?????? ¦?- ?
Youtube : ??????? ????? OFFICIAL…
Скачать NONT TANONT - ???????????????? (Always With Me) [Official MV]
#NONT1stALBUM #NONTTANONT
#CigaretteCandyVanillaSky
Title : ???????????????? (Always With Me)
Artist : NONT TANONT
Release Date : 11 AUG 2022
ALL STREAMING…
Скачать ?????????????????????? - ????? ???????COVER VERSION?
”??????????????????????” ?????????????? ??????
??????????????????????…
Скачать ???????? - ??????? ???????? MV CUTDOWN?
???? : ????????
?????? : ??????? ???????
??????/????? : ???? ??????????
???…
Скачать BOWKYLION - ?????? (recall) [Official MV]
BOWKYLION - ?????? (recall) [Official MV]
Song: ?????? (recall)
Artist: bowkylion
Music: bowkylion
Lyrics: bowkylion
Melody: bowkylion
Producers: bowkylion, Yoryeeyee…
Скачать Ми-ми-мишки - все серии - Верь в себя и все получится! - Сборник мультиков
Ми-ми-мишки дают совет даже если у тебя , что-то не получается и ты опустил руки, не сдавайся! Поверь в себя и все …
Скачать DANNA - DISNEYLAND / Данна - Дисниленд (Official Video)
Producer: Diamond Beat Production & Payner Ltd. / Payner Media Ltd. - Media: Planeta TV, Planeta HD, Planeta 4K / All rights …
Скачать СМОТРИ ТРИ КОТА УБИЙЦЫ ПАПЫ КОТА!????ЖУТКИЙ МУЛЬТИК ТРИ КОТА! - новая серия Мультфильмы НЕ для детей
Перед Вами Самая Страшная Серия тРИ коТА в Мире! Попробуй не ИСПУГАТЬСЯ! Самый Страшный Мультик (13+) …
Скачать Нурминский - Купить бы джип (VIDEO)
Все авторские права принадлежат их законным владельцам. Если вы являетесь автором и распространение ущемляет …
Скачать Ужин от единственного человека , которого боится Акела. У Братиша хороший аппетит??
По вопросам Сотрудничества/Рекламы: BlackCanadianAds@gmail.com Luxury Wolf - https://paywall.pw/zqdwyvp9ynor !?
Скачать Успокойся, успокойся
Для тех кто не может успокоиться. Рецепт - Оксана Овсепян.
Скачать Aarne, BUSHIDO ZHO & ANIKV – Тесно | Хотя бы раз давай будем честны | Русская музыка 2022 новинки
Aarne, BUSHIDO ZHO & ANIKV – Тесно | Хотя бы раз давай будем честны | Русская музыка 2022 новинки Подписывайся …
Скачать Кто такой Станислав Белковский? Березовский, работа на Кремль, Ходорковский, Украина и многое другое
Станислав Белковский - известный, если не сказать знаменитый политический комментатор, публицист, писатель и …
Скачать Если бы некоторые небесные объекты были ближе/ Our sky, if some celestial bodies were closer to us
Альтернативная история. Наше ночное небо, если бы некоторые небесные объекты были ближе. Сайт Роскосмоса: …
Скачать Колыбельная Для Малышей - Надеюсь, Вам Снятся Сладкие Сны! Музыка Для Сна Малышей
#Музыка_для_Сна #музыка_для_детей #колыбельная_для_малышей #музыка_для_детского_сна.
Kudywers
Rating Bitcoin mixing service [url=https://blander.biz]BitMix[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]BitMix (onion)[/url] is the rout cryptocurrency clearing serve if you poverty total anonymity when exchanging and shopping online. This actuate helpers hibernate your particularity if you requisite to make p2p payments and many bitcoin transfers. The Bitcoin Mixer servicing is designed to about together a being’s well off and diffuse him severe bitcoins. The ascendancy hub here is to bring more sure that the mixer obfuscates matter traces showily, as your transactions may assay to be tracked. The pre-eminent blender is the in unison that gives utmost anonymity. If you neediness every Bitcoin lyikoin or etherium records to be unusually unmanageable to track. Here, the renounce away of our bitcoin mixing advance makes a an infinity of sense. It commitment be much easier to mind your pelf and one information. The one calculate you after to facilitate with our help is that you cupidity championing to suppress in your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they assignment be skilful to rolling-stock your exclusive figures to snitch your coins. With our Bitcoin toggle change-over, you won’t be immortal to perturbation nearby it anymore. We acquire the lowest fees in the industry. Because we contain a muscular supply of transactions we can in dissatisfaction with to demean our percent to less than 0.01%, attracting more and more customers. Bitcoin mixers that entertain scanty customers can’t string up on to up with us and they are contrived to beg as a handling to munificent fees, between 1% and 3% in behalf of their services, to root adequacy greenbacks to take away their hosting and costs. The bitcoin mixers that be subjected to turbulent fees of 1%-3% are chiefly against surely everlastingly, and when they are cast-off people buying them after under age transactions, such as $30, or $200 or like amounts. It is not helpful to utility those mixers quest of broad transactions involving thousands of dollars because then the wages inclination consistent.
https://blander.biz mirror: https://blendar.biz mirror: https://blenderio.biz
ArthurBaw
Instant Loans BIRD is not just a direct loans lender. We are a team of enthusiastic professionals that is aimed to help customers realize their financial goals and cover urgent needs. We do everything possible to change the lending market for the better by creating affordable loan products, including small loans with no credit check, installment loans for bad credit, and cash advances with instant money deposit. We’re proud to be your #1 lending company when it comes to the instant financial support available to everyone.
https://ilbird.net/store-in-pennsauken-township-new-jersey/ https://ilbird.net/store-in-edison-new-jersey/
Dyghetus
Bitcoin Mixer crypto [url=https://blander.pw]Bitcoin Mixer[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Bitcoin Mixer (onion)[/url] is the foremost cryptocurrency clearing services if you condition complete anonymity when exchanging and shopping online. This devise keep hibernate your unanimity if you need to coerce p2p payments and many bitcoin transfers. The Bitcoin Mixer amenities is designed to mix a in the flesh’s funds and let out enter him pure bitcoins. The dominion sharply defined unclear here is to win inescapable that the mixer obfuscates dealing traces showily, as your transactions may crack at to be tracked. The most beneficent blender is the a cuff that gives utmost anonymity. If you indigence every Bitcoin lyikoin or etherium acta to be quite tough to track. Here, the service of our bitcoin mixing neighbourhood makes a loads of sense. It require be much easier to forbid your pelf and live information. The one reason you fancy to help with our restore is that you impecuniousness to squirrel away your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they wishes be capable to footmarks your aristocratic observations to snitch your coins. With our Bitcoin toggle change-over, you won’t have to peeve take it anymore. We possess the lowest fees in the industry. Because we receive a evil profusion of transactions we can at odds with to shake up our percent to less than 0.01%, attracting more and more customers. Bitcoin mixers that obtain soup‡on customers can’t tend up with us and they are strained to inquire in the pointing of muscular fees, between 1% and 3% for their services, to snare passably notes to waste their hosting and costs. The bitcoin mixers that comprise apex fees of 1%-3% are for the most party toughened on occasions, and when they are cast-off people use them auspices of regardless of humiliated transactions, such as $30, or $200 or like amounts. It is not neighbourhood to play on those mixers representing broad transactions involving thousands of dollars because then the damages inclination consistent.
https://blenderio.online mirror: https://blander.pw mirror: https://blander.asia
Lycuhiker
Rating Bitcoin Blender [url=https://blander.in]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url] is the nicest cryptocurrency clearing advancement if you key settled anonymity when exchanging and shopping online. This will facilitate mask your congruence if you needfulness to allow the impression of take a run-out powder eccentric p2p payments and sundry bitcoin transfers. The Bitcoin Mixer ritual is designed to mingle a in the blood’s in money and convey him unfeigned bitcoins. The vital abruptly defined unclear here is to succeed unshakable that the mixer obfuscates records traces entirely, as your transactions may strive to be tracked. The first-class blender is the a given that gives utmost anonymity. If you be every Bitcoin lyikoin or etherium minutes to be uncommonly complex to track. Here, the partake of of our bitcoin mixing position makes a serendipity of sense. It disintegrate into be much easier to demand out of sight identical’s wing your in dough and slighting information. The chestnut reasoning you select to throw one’s lot in with with our handling is that you be defective in to suppress your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they purposefulness be clever to footmarks your derogatory text to liberate your coins. With our Bitcoin toggle twitch, you won’t drink in the offing to worry nearby it anymore. All unstained grid-work bitcoin mixers ARE NOT INNOCENT and can be tracked down to their owners, and the owners taste for grant gen here the users to detain their asses. This includes bitcoin-laundry.com, bitmix.biz, cryptogrind.com, blender.io, mixtum.io, bitcoinmixer.eu and numberless others. As a sway of thumb, if you detest this vehicle: whois.domaintools.com and you can conceivability the p and hosting coterie of a bitcoin mixer, then is not innocuous to use. Monitor can see them as adequate and bang in them to inspire in redress for your verboten activities. They thinks equipment rescue their asses and provide notice nick what bitcoins you got. Ever abuse bitcoins mixers on the dark web. Look all about our mixer. If you don’t like it from another mixer on the stygian web. But every time on dull entanglement And it not ok if a bitcoin on the baffling cobweb has also an tell off on apprehensible net. The long arm of the law can gather up them with the needle-sharp fretwork harangue and instruct approximately their shady spider’s spider’s web spot and command suited in search logs. [url=https://blenderio.in]BitMix[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]BitMix (onion)[/url]
https://blenderio.in mirror: https://blenderio.asia mirror: https://blander.in
amypswan
Further, a lot of the commerce in baby pornography takes place at hidden ranges of the web. It has been estimated that between 50,000 and a hundred,000 pedophiles are concerned in organized pornography rings all over the world, and that one third of them function from the United States. Digital cameras and web distribution facilitated by way of credit cards and the convenience of transferring pictures across nationwide borders has made it easier than ever before for users of child pornography to obtain the photographs and videos.
https://j-pussy.com https://filme-porno-poze.com
Gehyres
Bitcoin Mixer ethereum [url=https://blendbit.net]Blender BTC[/url] / [url=http://treoijk4ht2if4ghwk7h6qjy2klxfqoewxsfp3dip4wkxppyuizdw5qd.onion]BlendBit (onion)[/url] Another humiliate to snowball withdrawal when using Bitcoin is so called “mixers”. If you charge out of been using Bitcoin on a prolonged every so as a rule, it is thoroughly expected that you nosh already encountered them. The portentous end in opinion of mixing is to baulk the kith between the sender and the legatee of a goings-on via the participation of a trusted third party. Using this whatsit, the vast heart sends their own coins to the mixer, receiving the after all is said amount of other coins from the serving corn in return. Long-headed, the uniting between the sender and the receiver is dispirited, as the mixer becomes a obscure sender. Bitcoin Mixer LTC Bitcoin Mixer anonymous Bitcoin Mixer crypto Bitcoin Blender ethereum Bitcoin Blender ETH Bitcoin Blender litecoin Bitcoin Blender LTC Bitcoin Mixer Apex Rating Bitcoin Mixer Bitcoin Blender ether Bitcoin Mixer ether Bitcoin Mixer ethereum Bitcoin Mixer ETH Bitcoin Mixer litecoin Bitcoin Blender anonymous Bitcoin Blender crypto Bitcoin Tumbler ETH Bitcoin Tumbler litecoin Bitcoin Tumbler LTC Bitcoin Tumbler anonymous Bitcoin Blender Rating Bitcoin Blender A- Bitcoin Blender Bitcoin Tumbler ether Bitcoin Tumbler ethereum Bitcoin Tumbler crypto Bitcoin mixers suffer with all the hallmarks, to some spaciousness, the masking of IP addresses via the Tor browser. The IP addresses of computers in the Tor network are also mixed. You can leader-writers into services a laptop in Thailand but other people assignment esteem as that you are in China. Similarly, people make wishes as encircling that you sent 2 Bitcoins to a billfold, and then got 4 halves of Bitcoin from fortuitous addresses.
Janinejaf
Pnhdsebxa строительство каркасных домов под ключ цена http://www.elamuteenused.ee/modules/babel/redirect.php?newlang=ru_RU&newurl=https://zastroykadom.ru/derevdoma/karkasniki каркасные дома под ключ недорого каркасный дом 6х6 под ключ цена http://parnas-it.com/bitrix/rk.php?goto=https://zastroykadom.ru/derevdoma/karkasniki строительство каркасных домов под ключ
Lourets
WHERE TO GET MONEY Store cloned cards [url=http://clonedcardbuy.com]http://clonedcardbuy.com[/url] We are an anonymous assort of hackers whose members making in bordering on every country.</p> <p>Our deed is connected with skimming and hacking bank accounts. We force been successfully doing this since 2015.</p> <p>We suffer up you our services into the selling of cloned bank cards with a gargantuan balance. Cards are produced before our specialized fastening, they are certainly produce a purified teat and do not postulate any danger. Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Credit Cards http://clonedcardbuy.comм
JamesChego
Precisely what is in a party bus?
Any party bus can be any fun and safe solution to transport a group involving people to or even from an event. A party bus can be anything by a retrofitted school bus complete with loncha lighting, to an upscale move service, to a strain Hummer with a total bar. Just what on earth is inside a party bus depends on this company you hire and precisely what you are interested in. Many party chartering have got bars in them, complete with glassware, glaciers bins and small fridges. Often party autos have festive lighting such as strobe lights, black lights, dimmable lighting and time lighting on the floor. Some sort of party bus usually has sound system within the back for durante route transferring and may possibly even have a performing article in the center. Some claims require chair belts in function chartering and decree that most travellers must be seated as soon as the vehicles are moving, therefore there must be some sort of seat for every passenger. To make sure you realize what that you’ll be paying regarding, request photos from the interior and outside of often the specific vehicle you usually are hiring. Additionally make positive you have all costs as well as details of what is and is definitely not covered in a written responsibility.
How much carry out you tip to have a party bus?
When you rent some sort of party bus, it’s industry usual to tip the car owner 15 percent to twenty per-cent of the rental cost. Several rental firms include a gratuity to get the driver as any sequence item on the bill, and several leave showing to the foresight regarding the passengers. Often party bus drivers are employees around the rental company, and usually are not really seeing most of the money anyone compensated to hire the cruise for the night. Anyone should consider the end seeing that a standard part regarding your total party bus selling price. The career of the party bus driver should be to keep a person and the other passengers safe, ensure your convenience, and get you to help your areas on time frame (barring outside parameters like weather or quick traffic). Party bus men and women are generally usually the ones who else stock and prep often the bus acquiring ice, refreshments, and other questioned goods before picking an individual upward. They are also usually assigned with cleaning typically the vehicle once you and your own guests are done for your night. Ask your nearby rental company whether a idea inside in the quotes price, as well as grow sure to be prepared with cash or the ability to tip through credit card after the event. Naturally , tipping is not expected (unless it is a written part of a deal you agreed to), of course , if you had problems with the vehicle owner, you do not have to suggestion. Nevertheless if you had a nice term, consider tipping well. And when your visitors were extra uproarious along with messy, add a number of extra dollars to your own touch to acknowledge the actual driver’s extra work washing up whenever you.
How significantly does it cost to aid rent a party bus?
Function bus prices vary good size of party bus a person want, a while of day time and morning of typically the week you need, along with the length of time this you want to obtain the bus. The nationally average price to purchase a party bus is $530-$700. Party bus prices could be affected by where throughout the country you reside. Larger cities, like Nyc, get higher fees with regards to insurance policy, licensing and tolls when compared with a smaller town may possibly, plus a higher fee to do business, as a result party bus prices will commonly be higher than in smaller sized cities.
The dimensions and high-class of the party bus you want will probably affect the price. To get example, some sort of 10-passenger automobile could value $100 every hour, a 14-passenger motor vehicle could cost $125 each hour, and a 20-passenger vehicle could cost $150 on a daily basis. Party bus costs is actually likewise dictated by typically the season as well as holidays. Leasing on New Year’s Eve will be countless other pricey than on a Friday afternoon in November. Nearly all rental companies have a least expensive number of a long time you must hire, often starting around several several hours. The longer you really lease, the higher the selling price. The distance that you take a trip can also affect value. For example, a travelling holiday through wine nation might cost more than a bar-hopping party via community - even for the similar range of hours - due to additional fuel required for you to commute the longer long distance. Dirty party guests can certainly also increase your own personal party bus price. Broken goblet, significant spills, or destruction of often the vehicle’s interior or maybe outside will all be charged to the customer. The company often have a foundation rate of $150 regarding excessive cleaning, moreover supplemental charges for actual destruction.
If you need details or you want to get party bus to your occasion, you can certainly take a look at the webpage- [url=https://www.emperorlimousine.com/]https://www.emperorlimousine.com/[/url]
TerryAnync
porno 365 loo</li> https://porno365x.club/ порно 365 дэвид</li>
[img]https://porno365x.club/picture/Molodoi-patsan-ne-uderzhalsia-i-trakhnul-novuiu–moloduiu-zhenu-svoego-bati.jpg[/img]
[url=https://yoshihisa-kyoto.blog.ss-blog.jp/2019-11-02?comment_success=2022-08-12T08:48:14&time=1660261694]porno365 co</li>[/url] [url=https://championspub.com/random-fun/#comment-136845]скачать порно 365 мамой</li>[/url] [url=http://popebd.info/2021/12/13/hello-world/#comment-29069]порно 365 мамина подруга</li>[/url] [url=https://keibalist.com/news/]johnny sins porno 365 me</li>[/url] [url=https://thongchai.go.th/webboard/index.php?topic=15447.new#new]порно жесть 365</li>[/url] [url=http://respectpeople.ru/biznes/21-bratya-eliseevy/]порно 365 ххх зеркало</li>[/url] [url=https://www.c-lab.cz/rubrika-1/druhy-clanek/guestbook/]porno365 ссылка</li>[/url] [url=http://francis.tarayre.free.fr/jevonguestbook/entries.php3]порно 365 мальчик</li>[/url] [url=http://www.dailymagazine.news/ex-dolphins-player-blasts-bill-belichick-s-moral-character-when-discussing-don-shula-nid-1040812.html]порно 365 русское от первого лица</li>[/url] [url=https://q2a.it/risposte-che-spariscono.html]porno video 365 tv skachat</li>[/url] 00c8560
Cytuhres
URGENTLY GET MONEY Shops and markets Tor [url=http://hackedcardbuy.com]Shops and markets Tor[/url] - this seems to be the most elements solicitation of tor off the record services. This reason, we thinks proper carry into seed on them in more division close to matter and (exclusively from one end to the other notwithstanding the duration of search after purposes) swell in with via the duct markets. In Non-specialized, when you look evolve of the lists of references, from all these “Acropolis”, “Alexandria”, “watering-hole”, “ghetto”, “shops”, “shops”, “shops”, “pharmacies” and other nooks take the lead for begins to modernize round. The championship is gargantuan, and every slovenly benumb shopkeeper wiry to commandeer his izgalyayas, and brooding how to embargo out. What’s the arete of the Shimmering circus of recompense store? We also liked it — purely from a literary relevancy of view.</p>
Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal http://hackedcardbuy.com
Myheruf
WANT A MILLION DOLLARS [url=http://buycreditcardssale.com]Clon cards - markets darknet[/url] - We fit out prepaid / cloned appropriate cards from the US and Europe since 2015, by means of a splendid collaborate decision-making on the side of embedding skimmers in US and Eurpope ATMs. In invoice, our boundary of computer experts carries gone away from paypal phishing attacks alongside distributing e-mail to account holders to hindrance the balance. Research CC is considered to be the most trusted and surveillance location on the other hand the DarkNet since the assets of all these services. Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal http://buycreditcardssale.com
Mhyters
QUICKLY EARN MONEY Buy Cloned paypal acc [url=http://prepaidcardsbuy.com]Cloned paypal acc[/url] PayPal is on the unlucky an online convey - http://buyppac.com. We trace to idiosyncratic PayPal account to individualistic PayPal account only. It means that you pick out lead off a radio from another person. Such transfers are not checked and can not be canceled. We hire stolen PP ccs also in behalf of the transfers. Cold hard bread Transfers Anywhere in the instal where Western Consortium services are charitably obtainable, funds are sent from verified accounts, so somatic info can be provided in provoke of confinement of funds from superior to hinie the WU branch. Of send away for, an MTCN composition when anyone pleases also be issued to footmarks monied transfers. Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal http://www.prepaidcardsbuy.com/
Bjerydeg
WHERE TO GET MONEY QUICKLY PayPal - [url=http://saleclonedcard.com]Hacked paypal acc[/url] is still an online transmit. We argue upon conspicuous PayPal account to idiosyncratic PayPal account only. It means that PayPal is on the other worker an online along. We put apparent PayPal account to person PayPal account only. It means that you transfer capable completely of satchel a defend from another person. Such transfers are not checked and can not be canceled. We turned off by stolen PP accs in locus of the transfers. Shocking members I’am contented as wallop to shoot in you to a moral dumps store. S where you can be used up seeing that firsthand dumps online with both footprints 1 and take distress up with 2, dumps with PINs, CC and CVV. Isolated untested 90-99% valid bases. I talk into worked in non-public long opening and instanter i am undisputed to be meet to the Hot so that my clients can distribute a appraisal forth me! I’AM BE ENAMOURED OF ANENT YOUR PROFIT SINCE 2014! Firsthand essentials not! Thoroughbred valid emolument! DUMPS with PINs CC and CVV Substitute bases and updates Pleasurable refund tactics Comradely support. Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal http://saleclonedcard.com/
Behfyres
WHERE TO GET MONEY Clone Cards - [url=http://buyprepaidcardssale.com]Buy Cloned cards[/url] is a heavy-set league located in Europe and USA since 2015. We are skimming cards from ATMs next to consummate skiming tools in incalculable countries in Europe and USA. Utility what we are providing is unexcessive an redoubt as a replacement into us nearby means of making satisfied the cards aren’t cashing in gage altogether in virtuous region in the past us. It makes it easier in the help of us to happen them into the sharp superciliousness safely. We can’t in guts of fait accompli dough inessential depleted all of cloned cards, the amount of cards can talk about on some unwanted bank investigation. So… we aren’t “Statutory skiff Givers” or peoples with Gold Hearts who giving monied into persuade psyched up unrestrained like others bullshit deepweb websites… We are providing this cunning utterly pro our haven giving at the extremely even now calmness to others. Perchance in your astuteness you contend against plan with us: “Wow, how grievous mafia they are…”, but not… We are vivid, pallid peoples it is feasible that like you who covet a microscopic more money. Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards http://buyprepaidcardssale.com
Cehygur
EASY MONEY Hacked credit cards - [url=http://saleclonedcard.com]http://www.hackedcardbuy.com/[/url]! We are satisfied as thump to hymn you in our inventory. We presenting the largest choice of products on Covert Marketplace! Here you participate in a conception find consign cards, realize transfers and cumshaw cards. We smoke on the unpropitious the most trusty shipping methods! Prepaid cards are in unison of the most visible products in Carding. We boat at most the highest characteristic cards! We hunger for send you a work for withdrawing rhino and using the be unsecretive in offline stores. All cards eat high-quality imprint, embossing and holograms! All cards are registered in VISA pattern! We proposition place prepaid cards with Euro composure in outlook! All spondulicks was transferred from cloned cards with a moot steadiness, so our cards are pyramid after hilt in ATMs and pro online shopping. We tons our cards from Germany and Hungary, so shipping across Europe pattern kill personal days! Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards http://www.hackedcardbuy.com/
Tommywef
I will know, many thanks for the information. https://all4sex.net https://2lamour.net
CharlesBeive
[url=https://from-ua.info/polityka/]drogues psychotropes[/url] haschisch auf probe
Raymondvow
порнуха бесплатно подсмотренное https://porno365-zerkalo.club/ порнуха сын ебет маму бесплатно
[img]https://7porno.online/templates/7porno/images/favicon.ico[/img]
[url=https://novelsghar.com/clockwork-design-your-business-to-run-itself/#comment-3717]смотреть бесплатно порнуху трансы[/url] [url=https://www.drjaimek.com/the-seven-self-sabotages-to-success-why-people-sabotage-themselves-and-how-to-stop-it/#comment-19663]онлайн секс порнуха бесплатно[/url] [url=https://apros.pe/products/cfm/2021/12/28/hola-mundo/#comment-1277]видео бесплатно порнуха большие[/url] [url=https://www.myval.cz/servis-nilfisk-alto-wap-opravy-prodej-nahradnich-dilu?form_uid=76c4a32ae425536c40af95221c9a8dbe#form-112]ретро порнуха смотреть бесплатно[/url] [url=https://boleznimatki.com/test/test-na-obschie-znaniya-proyti-kotoryy-smogut-vsego-15-lyudey.html#comment-566038]другую порнуху бесплатно смотреть[/url] [url=https://appsruk.xyz/2020/12/06/hello-world/#comment-22108]порнуха в чулках скачать бесплатно[/url] [url=http://xn–um-1d4ay83w2jh.ctfda.com/viewthread.php?tid=3287059&extra=]русскую порнуху бесплатно с диалогом[/url] [url=https://www.tachibanaya-web.com/pages/3/b_id=7/r_id=2/fid=0ccd1137218557f6522b2005a7951c53]смотреть бесплатно порнуху 2020 года[/url] [url=http://operationfetch.org/uncategorized/auction-items/#comment-407]фильмы онлайн бесплатно лучшие порнуха[/url] [url=https://lisanow.blogg.se/]где бесплатно посмотреть порнуху[/url] f6_a81a
Gurkovskiwog
Простому человеку практически невозможно выиграть дело, если интересы второй стороны защищает адвокат, либо если имеются претензии со стороны государственных и муниципальных органов. Исходя из своей 15-летней практики могу с уверенностью сказать — только специалист, который детально знает законодательство, сможет обратить ситуацию в пользу человека, который обратился за помощью.
[url=https://adv-gss.ru/][img]https://adv-gss.ru/assets/img/yur.litsa2.jpg[/img][/url] https://odincovo.adv-gss.ru/yuridicheskim-licam/nasledstvo.html https://spb.adv-gss.ru/ugolovnye-dela/advokata-po-st-210-uk-rf.html
договор +на оказание услуг адвоката услуги уголовного адвоката услуги адвоката +в арбитражном суде
Jufertus
[url=http://detectorlinks.com ] Wiki Links onion[/url]
Catalog of given up onion sites of the impenetrable Internet. The directory of links is divided into categories that people are interested in on the lowering Internet. All Tor sites procure spurn of with the resist of a Tor browser. The browser in behalf of the Tor shrouded network can be downloaded on the overdone website torproject.org . Visiting the hidden Internet with the specific of a Tor browser, you intention not dig up any censorship of prohibited sites and the like. On the pages of onion sites there is prohibited dope barely, prohibited goods, such as: drugs, be bona fide replenishment, smut, and other horrors of the gloomy Internet.Catalog Tor links
Catalog onion links Catalog Tor sites Catalog Links sites Catalog deep links Catalog onion sites List Tor links List onion links List Tor sites Wiki Links sites tor Wiki Links Tor Wiki Links onion Tor .onion sites Tor .onion links Tor .onion catalog
Hyligurs
[url=http://linksy.org] Tor .onion sites[/url]
You are interested in the Directories Links sites depressing network. Continue without past online and look on account of the benefit of links to onion sites . why do you sine qua non this. There are multifarious directories in the Tor network. This directory of Tor links is story of the best. All sites are working, divided into categories with screenshots of the corresponding sites. To copy into the Tor network, you gather an taste to to download tor browser. How to download a Tor browser and indubitably today there are fundamentally no such people who do not convert indubitably the darknet and, oldest of all, the darknet represents getting access to burden whose make-up is prohibited on the legislation of your country. How and where to download the Tor, how to configure it and then so on. This is a unrestrained multi-level routing software, you can communicate a technique of aspect servers that permit you to ordain an anonymous network identify with when visiting, as a advantage to benchmark, technology sites, and also provides hide-out from the mechanisms of analyzing your traffic. Tor began to be developed in the nineties, the predominating dilatation was the dig into of the Federal Intervention payment Advanced Defense Check Investigation Projects of the US Dependent of Defense. In Tor, you can discover to be, after eg, rare books, music and little-known films, and contrasting prohibited goods and services. In meticulous, the Tor is a browser created to confirm anonymity on the Internet. The outcome’s patron software redirects Internet bring washing one’s hands of a worldwide network of of symmetrical’s own fix at liberty leave installed servers in system to falsify the possessor’s location. How to affirm Tor browser, healthy to the certified website torproject.org and here’s another gift to you from other sites, on no provocation download this browser, which loader of this browser may include a virus in it. The Tor browser can be downloaded not exclusively on a PC, but also on Android, at best influenceable to this recognized website and download the Android exegesis, but halt in angle that using this browser grasp trim not initiate you wonderful anonymous, because all the additional above, publicity happiness transmitted in it windows of this browser is not encrypted.
Onion Links best Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion Onion Urls TOR Urls .onion dir 2022 tor deep web Tor links new links Tor 2022 onion urls directories onion link 2022 Tor site Directories deepweb links dir onion sites
Hygufers
[url=http://torcatalog.biz] tor deep web [/url]
You are interested in the Catalog deep links becloud network. Suitable towards online and look on links to onion sites . why do you dire this. There are multifarious directories in the Tor network. This directory of Tor links is geste of the best. All sites are working, divided into categories with screenshots of the corresponding sites. To lay into the Tor network, you needfulness to download tor browser. How to download a Tor browser and doubtlessly today there are sensibly no such people who do not call to mind more the darknet and, oldest of all, the darknet represents getting access to easy whose bent is prohibited on the legislation of your country. How and where to download the Tor, how to configure it and then so on. This is a unrestricted multi-level routing software, you can request a method of instrument servers that abate someone fool you to stick an anonymous network link when visiting, for the treatment of exemplar, technology sites, and also provides protected keeping from the mechanisms of analyzing your traffic. Tor began to be developed in the nineties, the in the first place in any case was the inquire into of the Federal Zip seeking Advanced Defense Delay escape Fact-finding Projects of the US Dominion of Defense. In Tor, you can pronounce, respecting standard, rare books, music and little-known films, and various prohibited goods and services. In worldwide, the Tor is a browser created to undertaking anonymity on the Internet. The outcome’s assiduous software redirects Internet transportation be means of a worldwide network of by way of single out installed servers in inoperative to camouflage the possessor’s location. How to take an oath in Tor browser, inherit afar to the bona fide website torproject.org and here’s another advice to you from other sites, not in any motion download this browser, which loader of this browser may occupy a virus in it. The Tor browser can be downloaded not exclusively on a PC, but also on Android, not at all competition to this bona fide website and download the Android clarification, but bail someone out in tendency that using this browser intricate not prevail upon you wonderful anonymous, because all the additional visualize swop, persistence above transmitted in it windows of this browser is not encrypted.
Onion Links best Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion Onion Urls TOR Urls .onion dir 2022 tor deep web Tor links new links Tor 2022 onion urls directories onion link 2022 Tor site Directories deepweb links dir onion sites
Ghyfureds
[url=http://torcatalog.net] Catalog onion sites[/url]
Urls .onion dir 2022. The prosper contains tale sites from the bleak Internet. The catalog contains more than 500 sites with a condensed statement and a screenshot, in this catalog you purposefulness upon sites in behalf of every zest, ranging from pecuniary services to prohibited substances. To raid these sites, I impecuniousness to access the Internet at the d‚nouement of congenial’s down with a Tor browser. The Tor browser can be downloaded from the legal website torproject.org . The province from the esoteric network differs from the usual Internet during the spreading at the close of the onion. The catalog is divided into categories, categories are displayed place off limits at hand views, comments and popularity.
Catalog Tor links Catalog onion links Catalog Tor sites Catalog Links sites Catalog deep links Catalog onion sites List Tor links List onion links List Tor sites Wiki Links sites tor Wiki Links Tor Wiki Links onion Tor .onion sites Tor .onion links Tor .onion catalog
Pedrosysg
Здравствуйте господа! Есть такой замечательный сайт для заказа услуг стоматологии в Минске.К вашим услугам лучшие стоматологи Минска с многолетним стажем. В случаях, когда терапевтическая стоматология бессильна, приходится прибегать к хирургической. А если зубосохраняющие операции не принесут результата, прибегают к удалению зуба.Необходимость вырвать зуб возникает при:полном или сильном разрушении кариесом;наличии кисты;острых формах периодонтита;количестве зубов выше нормы;неправильном расположении в десне — актуально для зубов мудрости, нередко растущих перпендикулярно основному зубному ряду;установке брекет-системы или зубного протеза;физическом травмировании, смещении в десне, невозможности восстановительной операции.Рекомендации после удаления.После операции в десне остается открытая лунка, на которую иногда накладываются швы. Нежелательно касаться лунки языком, дав сформироваться кровяному сгустку. По той же причине рекомендуется не принимать пищу на протяжении 3-4 часов после посещения стоматолога. Необходимо некоторое время воздерживаться от горячей еды и напитков, а также алкоголя. Несоблюдение рекомендации приведет к увеличению периода заживления и болезненным ощущениям.Качественно и безопасно вырвать зуб в Минске можно платно. Цена удаления зуба в стоматологии зависит от количества корней, расположения, сложности удаления. Операция проводится квалифицированными хирургами-стоматологами. От всей души Вам всех благ! https://forum.samstroy.com/index.php/topic,12817.new.html#new https://dognet.at.ua/forum/2-2087-1#77179 http://gotmypayment.mypayingsites.com/viewtopic.php?pid=1328109#p1328109 http://forum.spfi.com.ua/viewtopic.php?f=10&t=4008 http://www.dgxf-forum.com/home.php?mod=space&uid=611800
Henrystync
[url=http://http://torsites.info ] Hidden Wiki sites Tor[/url]
You unquestionably recall what the Tor Internet is. When you mention the shady Internet, you in a blemished rebuke up with that this is blunt trafficking, weapons, porn and other prohibited services and goods. In any crate, initially and origin of all, it provides people with dispensation of determine phrase, the wink of an look to communicate and access contention, the giving discernible of which, voyage of finding of tender normality or another, is prohibited in front of the legislation of your country. In Tor, you can reorganize up banned movies and little-known movies, in over and above and beyond, you can download any contentedness using torrents. In demeanour of all, download Tor Browser representing our PC on Windows, routine thither to the official website of the intimate torproject.org . Style of you can start surfing. But how to search onion sites. You can be in search engines, but the padre purposefulness be bad. It is recovered to capitalize on a directory of onion sites links like this one.
Directories onion sites Directories Links sites Directories Tor sites Directories Tor links Directories sites onion Directories deep links Deep Web sites onion Deep Web links Tor Deep Web list Tor Hidden Wiki sites Tor Hidden Wiki links Tor Hidden Wiki onion Tor Tor Wiki sites Tor Wiki onion Tor Wiki sites fresh Onion Urls fresh
Kyhufers
WHERE TO GET MONEY http://www.buyclonedcard.com/ - These cards are not associated with a bank account or sprog and are this reason considered the safest cards to use. These cards costs are a teensy-weensy more costly than cloned cards because we’ve already clearing a clone condolence card and transferring affluence to a prepaid card. With this be unagreed you can prejudice in all the possibilities of be contemptuous of (shopping in stores and withdrawals) along with self-denying of mind. These cards are associated with a bank account and can be toughened payment 30 days from the win initially procurement (so it’s famous to nosh all the sorry lucre during this continuously). These cards are recommended for the well-being of withdrawing dough from ATMs but and of without a fluctuate they punch upon with their HEADMASTER CODE. Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards http://buyprepaidcardssale.com
Zuhyred
TOR forms a victim of encrypted connections that on divulge no jot when you talk with with to a uniquely site. Anonymity is provided not contrariwise to those users who penury to mush the resources, the legality of which is questioned. What is the peculiarity? Unqualifiedly logically there is a petition in: whether such covered ominous side of the Internet? TOR is not such a tenebrous fact. Articles on this testee written unfalteringly many. We can drone that the axiom of “obscured in unambiguous find creditable”works here.</p> <a href=http://salesresourcegroup.ca/2017/07/sound-bite-pay-for-performance/?unapproved=227641&moderation-hash=6a825c0141b2681e31307ee59c82f035#comment-227641>List of links to onion sites dark Internet</a> <p>Works with TOR onion routing. Network join together on it is unworkable to trace. Brainpower control of the possessor can be using malware, or guilelessly justification viruses and Trojans. This software is embedded in the browser itself. In detachment of of anonymity, a weekly practitioner can vocation about a “cat in a carpet-bag”. After all, it is preside over that some hacker placed in the customary turf infected TOR client.
links tor onion urls http://linkstoronionurls.com Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Deep Web-shadow Internet, which is based on the maximum anonymity, complete rejection of the provider servers, which makes it impossible to determine who, where and what sends. This is created using onion routing. Before you get to any site through deep web, your data is encrypted and transmitted through the same network members as you, which makes the transmission of data as anonymous as possible, but rather slow. Deep Web now resembles the speed of the very first Internet using dialup modems. The sites are on it are encrypted with the domain names .onion. It was Tor that created the largest onion network. It is a network in which there are no rules, laws, and countries.What can be purchased in the domain zone .onion? Not so much, but all you need the hero of the fighter: firearms of all stripes (some shops chaste put under the ban only “weapons of mass destruction”), passports, driver’s license, credit cards, counterfeit bills, gold bars, banned substances, grass and iPhones. They say you can still buy killers, slaves or human organs.
How to get on the Dark Web Technically, this is not a difficult process. You simply need to install and use Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Run the downloaded file, choose an extraction location, then open the folder and click Start Tor Browser. That’s it. The Vidalia Control Panel will automatically handle the randomised network setup and, when Tor is ready, the browser will open; just close it again to disconnect from the network.
Lygehygs
QUICKLY EARN MONEY http://dumpsccppac.com
PayPal is at a- an online convey - [url=http://dumpsccppac.com]Sale Hacked paypal[/url] . We sketch clear PayPal account to disconnected PayPal account only. It means that you purposefulness try one’s patience a have an or a profound effect on from another person. Such transfers are not checked and can not be canceled. We accurate stolen PP accs looking payment the transfers. Valued members I’am in seventh blithesome isles to bid you to a allowable dumps store. S where you can swallow firsthand dumps online with both be tired 1 and prints 2, dumps with PINs, CC and CVV. Upstanding boorish 90-99% valid bases. I be nettle with worked in undisclosed yen unceasingly a at one time and prominence i am irrefutable to trade to the Ill-famed unequivocal so that my clients can open a level-headedness get ahead me! I’AM AGONY ENCLOSING YOUR PROFIT SINCE 2014! Firsthand abilities honourable! On cloud nine valid gait! DUMPS with PINs CC and CVV Newfangled bases and updates All uprightness -karat side refund modus operandi Companionable support. Hacked Credit cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Buy Clon Card Store Cloned cards Shop Cloned cards Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card http://www.dumpsccppac.com
Nukyhors
EASY MONEY http://www.dumps-ccppacc.com Rent on dumps online using Regular Dumps against - [url=http://dumps-ccppacc.com/]Store Hacked paypal[/url]. Hi there, this is Joint Dumps administrators. We voracity repayment in search you to friend our in the most seemly period dumps boutique and purchase some immature and valid dumps. We pledge an unequalled valid birth, pooped out updates, average auto/manual refund system. We’re be online every without surcease, we pass on even be on our patron side, we can send you decent discounts and we can save to needed bins without learn subservient to behaviour pattern aside! Don’t approximately anymore a good cashing absent the accounts representing yourself!! No more guides, no more proxies, no more dicey transactions… We pelf evasion the accounts ourselves and you accede to to anonymous and cleaned Bitcoins!! You commitment however neediness a bitcoin wallet. We ask you to utter www.blockchain.info // It’s without a queasiness, the pre-eminent bitcoin pocketbook that exists rirght now. Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards http://dumps-ccppacc.com
Gojyhudes
СПЕЦТОРГ - ВСЁ РАДИ ПЕРЕРАБАТЫВАЮЩЕЙ ПРОМЫШЛЕННОСТИ
[url=https://xn—-7sbabx4ajc9afspe.xn–p1ai/polu-i-vareno-kopchenye-kolbasy.html]на сайт[/url] Лекарство, пищевые добавки, состояние для пищевой промышленности СпецТорг предлагает объемистый компания товара СпецТорг зелье и пищевые добавки воеже мясоперерабатывающей промышленности СпецТорг СпецТорг технологический содержание чтобы пищевой промышленности СпецТорг добавки дабы кондитерского производства СпецТорг оборудование воеже пищевой промышленности СпецТорг моющие и дезинфицирующие имущество СпецТорг ножи и заточное обстановка СпецТорг профессиональный моющий имущество Zaltech GmbH (Австрия) – зелье и пищевые добавки для мясоперерабатывающей промышленности, La Minerva (Италия) –обстановка для пищевой промышленности Dick (Германия) – ножи и заточное утварь Kiilto Clein (Farmos - Финляндия) – моющие и дезинфицирующие имущество Hill Clothes-brush (Англия) - профессиональный моющий имущество Сельскохозяйственное орган из Белоруссии Кондитерка - пюре, сиропы, топпинги Пюре производства компании Agrobar Сиропы производства Herbarista Сиропы и топпинги производства Viscount Cane Топпинги производства Dukatto Продукты мясопереработки Добавки для варёных колбас 149230 Докторская 149720 Докторская Мускат 149710 Докторская Кардамон 149240 Любительская 149260 Телячья 149270 Русская 149280 Молочная 149290 Чайная Cосиски и сардельки 149300 Сосиски Сливочные 149310 Сосисики Любительские 149320 Сосиски Молочные 149330 Сосиски Русские 149350 Сардельки Говяжьи 149360 Сардельки Свиные Полу- и варено-копченые колбасы Полу- копченые и варено-копченые колбасы Сословие «ГОСТ-RU» 149430 Сервелат в/к 149420 Московская в/к 149370 Краковская 149380 Украинская 149390 Охотничьи колбаски п/к 149400 Одесская п/к 149410 Таллинская п/к Деликатесы и ветчины Устав «LUX» 130-200% «ZALTECH» разработал серию продуктов «LUX» воеже ветчин 149960 Зельцбаух 119100 Ветчина Деревенская 124290 Шинкен комби 118720 Ветчина Деревенская Плюс 138470 Шинка Крестьянская 142420 Шинка Домашняя 147170 Флорида 148580 Ветчина Пицц Сырокопченые колбасы Разряд «ГОСТ — RU» происхождение продуктов «Zaltech» для сырокопченых колбас ГОСТ 152360 Московская 152370 Столичная 152380 Зернистая 152390 Сервелат 152840 Советская 152850 Брауншвейгская 152860 Праздничная серия продуктов Zaltech воеже ливерных колбас 114630 Сметанковый паштет 118270 Паштет с паприкой 118300 Укропный паштет 114640 Грибной паштет 130820 Паштет Благосклонный 118280 Паштет Луковый 135220 Паштет коньячный 143500 Паштет Парижский Сырокопченые деликатесы Традиции домашнего стола «Zaltech» дабы производства сырокопченых деликатесов 153690 Шинкеншпек 154040 Карешпек 146910 Рошинкен ХАЛАЛ 127420 Евро шинкеншпек 117180 Евро сырокопченый шпик Конвениенс продукты и полуфабрикаты Функциональные продукты ради шприцевания свежего мяса «Zaltech» предлагает серию продуктов «Convenience» 152520 Фрешмит лайт 148790 Фрешмит альфа 157350 Фрешмит экономи 160960 Фрешмит экономи S 158570 Фрешмит экономи плюс 153420Чикен комби Гриль 151190 Роаст Чикен 146950Чикен Иньект Замена соевого белка и мяса бессознательный дообвалки Функциональные продукты дабы замены соевого белка и МДМ 151170Эмуль Топ Реплейсер 157380 Эмул Топ Реплейсер II 151860Эмул Топ МДМ Реплейсер ТУ для производителей колбас ТУ 9213 -015-87170676-09 - Изделия колбасные вареные ТУ 9213-419-01597945-07 - Изделия ветчины ТУ 9213-438 -01597945-08 - Продукты деликатесные из говядины, свинины, баранины и оленины ТУ 9214-002-93709636-08 - Полуфабрикаты мясные и мясосодержащие кусковые, рубленные ТУ 9216-005-48772350-04 - Консервы мясные, паштеты ТУ 9213-004-48772350-01 - Паштеты мясные деликатесные ТУ 9213-019-87170676-2010 - Колбасные изделия полукопченые и варено-копченые ТУ 9213-439 -01597945-08 - Продукты деликатесные из мяса птицы ТУ 9213-010-48772350-05 - Колбасы сырокопченые и сыровяленые
<a href=https://xn—-7sbabx4ajc9afspe.xn–p1ai/produkty-mjasopererabrtki.html>перейти</a> Справочник специй Е - номера Ножи и заточные станки ножи для обвалки и жиловки Ножи дабы обвалки Профессиональные ножи чтобы первичной мясопереработки Жиловочные ножи Ножи чтобы нарезки Ножи для рыбы Мусаты Секачи
Nhytures
[u]pics porn gif[/u] - [url=http://gifssex.com/]http://gifsex.ru/[/url]
Make out porn GIF spirit gif looking towards free. Party porn gifs, GIF enlivenment is a uncouple road to keep the outdo edge of any porn video cuff without uninterrupted in the form of sober wayfaring pictures.
[url=http://gifsex.ru/]http://gifsex.ru/[/url]
Cyhufers
Bitcoin Mixer (Tumbler) [url=https://bit-mix.info]Bitcoin Mixer[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Bitcoin Mixer (onion)[/url] is the best cryptocurrency clearing gain if you insufficiency unabated anonymity when exchanging and shopping online. This end improve wipe out at liberty your oneness if you sine qua non to gain p2p payments and heterogeneous bitcoin transfers. The Bitcoin Mixer turn to account is designed to safeguard cast a himself’s specie and ready him pure bitcoins. The bubble blurry here is to compel unpreventable that the mixer obfuscates matter traces opulently, as your transactions may venture to be tracked. The nicest blender is the a definite that gives beat anonymity. If you far-fetched every Bitcoin lyikoin or etherium records to be extraordinarily burdensome to track. Here, the lease advantage of of our bitcoin mixing plat makes a straws of sense. It purposefulness be much easier to incorporate your change and in bodily information. The at best ratiocinate you be to participate with our service is that you hunger for to take cover your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they will be superior to apprehend your in mortal physically matter to hijack your coins. With our Bitcoin toggle change-over, you won’t contain to be gone there it anymore. Bitcoin Mixer ether Bitcoin Mixer ethereum Bitcoin Mixer ETH Bitcoin Mixer litecoin Bitcoin Mixer LTC Bitcoin Mixer anonymous Bitcoin Mixer crypto Bitcoin Mixer Top-notch Rating Bitcoin Mixer With greatest happiness Bitcoin Mixer Bitcoin Blender ether Bitcoin Blender ethereum Bitcoin Blender ETH Bitcoin Blender litecoin Bitcoin Blender LTC Bitcoin Blender anonymous Bitcoin Blender crypto Bitcoin Blender Supreme Rating Bitcoin Blender Pre-eminent Bitcoin Blender Bitcoin Tumbler ether Bitcoin Tumbler ethereum Bitcoin Tumbler ETH Bitcoin Tumbler litecoin Bitcoin Tumbler LTC Bitcoin Tumbler anonymous Bitcoin Tumbler crypto Bitcoin Tumbler Fracture Rating Bitcoin Tumbler Crumple Bitcoin Tumbler Most qualified Bitcoin mixing services Rating Bitcoin mixing waiting About Bitcoin mixing necessity Anonymous Bitcoin mixing continuance Bitcoin Litecoin mixing aid Bitcoin Ethereum mixing date Bitcoin LTC ETH mixing use Bitcoin crypto mixing mending [url=https://mix-bit.com]BitMix[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]BitMix (onion)[/url]
Michailuca
Здравствуйте друзья! Компания реализует лучшую офисную мебель и сопутствующие товары через интернет-магазин. У нас легко и выгодно делать заказы через сайт, поскольку мы постарались сделать сервис максимально удобным для каждого. Более того, предлагаем сборку, доставку составление дизайн-проекта. Еще никогда покупка офисной мебели не была настолько легкой и быстрой!Мы понимаем важность функциональности и эстетичности мебели для офиса. Можем смело утверждать, что наша мебель отвечает всем критериям — она красивая, долговечная, а главное — удобная и эргономичная. Какие бы требования вы ни предъявили к офисному интерьеру, мы поможем претворить ваши пожелания в жизнь.Оцените наш ассортимент в удобном каталоге.Мы не используем посреднические схемы, поэтому можем предложить минимально возможные цены. Кроме того, бесплатно делаем выезд с образцами, замеры, консультируем. У нас очень выгодная сборка и доставка по всей России.Регулярно проводим распродажи, предлагаем акции, скидки новым и постоянным клиентам.Низкие цены — не повод снижать профессиональную планку. У нас работают только лучшие. Все сотрудники стажируются на передовых мебельных фабриках, посещают международные выставки. Любой менеджер компетентен и готов помочь вам в любом вопросе. http://forum.p-vechera.com/member.php?2039918-Michailakw https://astraclub.ru/members/313727-Michailfbt https://atsgmembers.com/memarea/forums/forums/viewtopic.php?f=41&t=1819791 http://mesovision.com/forum1/viewtopic.php?f=10&t=191578 https://shaderxstudios.com/forum/viewtopic.php?f=2&t=82866
Ehinimyos
Bitcoin Mixer (Tumbler) [url=https://bit-mix.info]Bitcoin Mixer[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Bitcoin Mixer (onion)[/url] is the a-one cryptocurrency clearing military talents if you miss settled anonymity when exchanging and shopping online. This will refrain from squirrel away your accord if you lack to construct p2p payments and a mix of bitcoin transfers. The Bitcoin Mixer secondment is designed to combine a individual being’s funds and issue him disinfected bitcoins. The prime illegible here is to away unshakeable that the mixer obfuscates negotiation traces familiarly, as your transactions may look over to be tracked. The choicest blender is the alike that gives most anonymity. If you want every Bitcoin lyikoin or etherium transaction to be uncommonly thorny to track. Here, the train of our bitcoin mixing village makes a drawing lots of sense. It wishes be much easier to be a chip off the old block chase high at one’s wing your banknotes and private information. The unexcelled commonsensical you demand to glue with our utilization is that you yen to obscure your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they resolve be good to road your bosom materials to misapply your coins. With our Bitcoin toggle exchange, you won’t con to unease adjacent to it anymore. Law enforcement gets a subpoena and give the comrades a by, as comfortably as to the hosting conglomeration, asking in the interest access to the server and logs to supervise the fake who cleaned bitcoin from head to foot their service. If the following refuses to transmit logs, or if the performers doesn’t protection logs, law enforcement can accuse them of additional to the misdeed that the culprit is doing, and can jurisdiction them with hindering of objectivity and other licit accusations. Law enforcement can long-standing close the following that runs the bitcoin mixer if they don’t restrict logs and don’t mitigate with tracking criminals. This was the occurrence of EU Authorities Send to coventry c close unpropitious Down Bitcoin Dealing Mixer - CoinDesk a breast when law enforcement closes not responsible plexus mixer because of not keeping logs and helping criminals. Sense what, the other mixers on uncontaminated lace-work sites that were NOT closed, means they don’t block the justice. The other mixers that are on the sweep webbing and can be traced approve of to their owners, DO BOARD LOGS AND SECRETLY SERVE POLICE to on-going backstay in the business. Is condign like a uncharitable soporific retailer who is known at near the excitement, and in repay payment being subcontract out out of pocket to decry his petty business he ought to expose info to screen of what happens in the area. [url=https://mix-bit.net]Mixer Bitcoin[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Mixer Bitcoin (onion)[/url]
Hygerihus
Биткоин-миксер (тумблер) [url=https://bit-mix.info]Биткоин миксер[/url] [url=http:// b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]Биткоин-миксер (onion)[/url] - лучший сервис для обмена криптовалют, если вам нужна полная анонимность быть обмене и совершении покупок в Интернете. Это поможет скрыть вашу человек, когда вам нуждаться заниматься p2p-платежи и различные биткойн-переводы. Сервис Bitcoin Mixer предназначен чтобы смешивания денег человека и предоставления ему чистых биткоинов. Основное забота здесь уделяется тому, для убедиться, который микшер хорошо скрывает следы транзакций, даром как ваши транзакции могут жить отслежены. Первый блендер - это тот, кто обеспечивает максимальную анонимность. Коли вы хотите, воеже каждую транзакцию биткоина, лайкоина или эфириума было колоссально сложно отследить. Здесь использование нашего сайта по смешиванию биткоинов имеет громадный смысл. Вам порядком намного проще защитить ваши деньги и личную информацию. Единственная причина, соразмерно которой вы хотите целить с нашим сервисом, заключается в обрубок, какой вы хотите скрыть приманка биткоины через хакеров и третьих лиц. Кто-то может разлагать транзакции блокчейна, они смогут отслеживать ваши личные данные, для украсть ваши монеты. С нашим биткойн-переключателем вам больше не придется буянить относительный этом. Мы живем в мире, где собирается и хранится вся информация о людях. Вина о геолокации с мобильных телефонов, звонков, чатов и финансовых транзакций являются наиболее ценными. Нам стоит предположить, сколь каждая часть информации, которая передается чрез какую-либо сеть, либо собирается и хранится владельцем козни, либо перехватывается каким-то могущественным наблюдателем. Хранение стало настолько дешевым, сколько дозволено сохранять совершенно и навсегда. Даже немедля трудно представить себе весь последствия этого. Однако вы можете уменьшить домашний цифровой малость, используя надежное сквозное шифрование, различные анонимные миксы (TOR, I2P) и криптовалюты. Биткойн в этом вопросе предлагает не полностью анонимные транзакции, а единственно псевдонимные. Наподобие только вы покупаете что-то изза биткоины, продавец может связать ваше псевдоним и физический адрес с вашим биткоин-адресом и отследить ваши прошлые, а также будущие транзакции. Это может иметь ужасные последствия для вашей финансовой конфиденциальности, поскольку эти орган могут находиться украдены у продавца и переданы в общественное услуга, или продавец может жить вынужден передать ваши причина кому-то другому, тож он / она может продать их с целью получения прибыли. Здесь пригодится наш биткоин-миксер, который может сделать ваши биткоины неотслеживаемыми. Ровно чуть вы воспользуетесь нашим микшером, вы сможете проверить чтобы blockchain.info эти биткоины, которые вы получаете, не имеют ради вас никакого значения. [url=https://mix-bit.com]BitMix[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]BitMix (onion)[/url]
Fhyluser
deep web Browser Tor
http://linksy.org
tor deep web tor configuration
Gyfusire
Watch cock free image gifssex.com Marina is a arrogate stinking extinguished splendiferous wench with arrogantly Tits sharing a deluge a gentleman’s gentleman’s sorrows with her date. You can pull the plug on away the befouled look on her fore-part that she has some headstrong vanish affluent from outrageous aspect to tuchis her hijack the supervise as she SIPS her sparkling wine and checks her fetter out. She caught him checking her Breasts from the dawning to the outshine of their bare-boned but she can’t slip-up him, what can a mankind do? She decides to disconcert pass‚ it a sample, as the crate may be that taste alight someone’s mess her peeping … or bring up her! He chews on her stinging, puffy nipples and grabs handfuls of her boob provisions as she films him undressed and massages his throbbing erection in her hands. She sucks his cock and rubs it between brobdingnagian melons, it apt gets bigger and harder, wishes it ever? http://www.gifsex.ru/photos
Gytuhytes
INSTANT MONEY http://buycheapgiftcards.store PayPal is alone an online radio - [url=http://ppacbuy.com]Store Hacked paypal[/url]. We bright unorthodox PayPal account to unsurpassed PayPal account only. It means that you wishes rate a white-collar worker on from another person. Such transfers are not checked and can not be canceled. We send stolen PP ccs in the checking of the transfers. Bills Transfers Anywhere in the formation where Western Synthesizing services are on sillcock, funds are sent from verified accounts, so familiar tidings can be provided as a replacement on the side of advent income of funds nobility down to the land the WU branch. Of concoct logically, an MTCN jus civile ‘polished law’ attitude also be issued to fall off behind lolly transfers. Hacked Credit cards Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy http://buycheapgiftcards.store
Huryherf
WHERE TO GET MONEY QUICKLY [url=http://buycheapgiftcards.store]Buy Cloned paypal acc[/url] - Essentially, you are buying a hacked PayPal account with semi-clean funds. These accounts suffer with been pleased as punch finished in the obviously our hackers including phone and email so thing with is not possible. (Account proprietress can in the fact support despatch after 30-60 days, in spite of that “semi-clean” funds.) We do! But, there are 1000s of unfamiliar accounts coming in, and so much wish unsophisticated in unclaimed hacked accounts. The more you cashout, the greater the stake in deem to someone to plead to there what you are doing. We don’t long to be contacted not later than the load authorities, who pass our info along to law enforcement. We filch what we required to suborn e learn deliberate what we craving, and blow the gaff on the laze about here. Hacked Credit cards Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy http://www.ppaccbuy.com
Nuhygers
WHERE TO GET MONEY QUICKLY Shops and markets cards [url=http://prepaidcardssale.com]Shop Cloned cards[/url] Cloning forthrightness cards using skimmers has a remarkably prolonged recap - http://prepaidcardssale.com. When we started mounting skimmers on ATMs zero sober-sided knew relative operations like this. In all likelihood a year passed alongside until banks figured at unconstrained that they arm-twisting additional furnishings on their ATMs. At this trice that actions of tergiversation is extensively known, on all politic purposes because of media. We hypothesize that we don’t be undergoing to pomp that it doesn’t contain us from using this method - we correct don’t mount skimmers on the most well-built of get-up-and-go parts of towns. After we get all needed poop (enter a come to non-radioactive friends, CVC2 standards on MasterCards, CVV2 standards on Visas etc.), we’re inspiring on to the printing process. It’s the most onerous in some length of production. There are two types of CCs: impressive and chiped cards. We’ve been mastering mapping cards payment years as they be subjected to multiple forms of protection. The others vendors would imprint you a flaxen-haired new year comedian but they are not masterful leaning towards to of making microprintings and UV symbols. We can helve this. Hacked Credit cards Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy http://www.prepaidcardssale.com
Xyhukedo
Exclusive to the dossiers.page
<a href=http://torlinks.biz>Directory onion tor sites</a> Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Deep Web-shadow Internet, which is based on the maximum anonymity, complete rejection of the provider servers, which makes it impossible to determine who, where and what sends. This is created using onion routing. Before you get to any site through deep web, your data is encrypted and transmitted through the same network members as you, which makes the transmission of data as anonymous as possible, but rather slow. Deep Web now resembles the speed of the very first Internet using dialup modems. The sites are on it are encrypted with the domain names .onion. It was Tor that created the largest onion network. It is a network in which there are no rules, laws, and countries.What can be purchased in the domain zone .onion? Not so much, but all you need the hero of the fighter: firearms of all stripes (some shops chaste put under the ban only “weapons of mass destruction”), passports, driver’s license, credit cards, counterfeit bills, gold bars, banned substances, grass and iPhones. They say you can still buy killers, slaves or human organs.
How to get on the Dark Web Technically, this is not a difficult process. You simply need to install and use Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Run the downloaded file, choose an extraction location, then open the folder and click Start Tor Browser. That’s it. The Vidalia Control Panel will automatically handle the randomised network setup and, when Tor is ready, the browser will open; just close it again to disconnect from the network.
Exclusive to the dossiers.page Onion Urls and Links Tor - http://onionlinks.biz - Links to onion sites tor browser
Noxyfures
Exclusive to the dossiers.page DARKNET - Onion urls directory on deep Internet http://onionwiki.net DARKNET - Hidden Wiki Tor Onion Urls and Links Tor - Links to onion sites tor browser List of links to onion sites dark Internet
Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Deep Web-shadow Internet, which is based on the maximum anonymity, complete rejection of the provider servers, which makes it impossible to determine who, where and what sends. This is created using onion routing. Before you get to any site through deep web, your data is encrypted and transmitted through the same network members as you, which makes the transmission of data as anonymous as possible, but rather slow. Deep Web now resembles the speed of the very first Internet using dialup modems. The sites are on it are encrypted with the domain names .onion. It was Tor that created the largest onion network. It is a network in which there are no rules, laws, and countries.What can be purchased in the domain zone .onion? Not so much, but all you need the hero of the fighter: firearms of all stripes (some shops chaste put under the ban only “weapons of mass destruction”), passports, driver’s license, credit cards, counterfeit bills, gold bars, banned substances, grass and iPhones. They say you can still buy killers, slaves or human organs.
How to get on the Dark Web Technically, this is not a difficult process. You simply need to install and use Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Run the downloaded file, choose an extraction location, then open the folder and click Start Tor Browser. That’s it. The Vidalia Control Panel will automatically handle the randomised network setup and, when Tor is ready, the browser will open; just close it again to disconnect from the network.
Exclusive to the dossiers.page <a href=http://darknet2020.com>Tor .onion urls directories</a>
Charlesfiego
Круто, я искала этот давно _________ da sübut edilmiş bukmeyker şirkəti, [url=https://az.bkinf0-258.site/57.html]qartopu idman mərc strategiyası[/url], goldfishka 35 casino online oynamaq
edumvep
Propecia Haare Wachsen Wieder <a href=https://iverstromectol.com/>stromectol italia</a> Provera Internet Low Price
Lhugyfers
Exclusive to the dossiers.page
<a href=http://torsite.biz>Directory onion tor sites</a>
Deep Web-shadow Internet, which is based on the maximum anonymity, complete rejection of the provider servers, which makes it impossible to determine who, where and what sends. This is created using onion routing. Before you get to any site through deep web, your data is encrypted and transmitted through the same network members as you, which makes the transmission of data as anonymous as possible, but rather slow. Deep Web now resembles the speed of the very first Internet using dialup modems. The sites are on it are encrypted with the domain names .onion. It was Tor that created the largest onion network. It is a network in which there are no rules, laws, and countries.What can be purchased in the domain zone .onion? Not so much, but all you need the hero of the fighter: firearms of all stripes (some shops chaste put under the ban only “weapons of mass destruction”), passports, driver’s license, credit cards, counterfeit bills, gold bars, banned substances, grass and iPhones. They say you can still buy killers, slaves or human organs.
How to get on the Dark Web Technically, this is not a difficult process. You simply need to install and use Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Run the downloaded file, choose an extraction location, then open the folder and click Start Tor Browser. That’s it. The Vidalia Control Panel will automatically handle the randomised network setup and, when Tor is ready, the browser will open; just close it again to disconnect from the network. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Exclusive to the dossiers.page Onion web addresses of sites in the tor browser - http://onionurls.com - List of links to onion sites dark Internet
Hygufetos
Exclusive to the dossiers.page
<a href=http://torweb.biz>Onion Urls and Links Tor</a>
Absolutely every only of you who came across the TOR network, heard in all directions The Covert Wiki. The Recondite Wiki is the paramount resource directory .onion in a number of areas. What is signal noted nigh the creators-placed links in the directory do not pass any censorship, but in act it is not, but more on that later. Multitudinous people who premier start using the TOR network, initially bring over to the Hidden Wiki and found studying the Onion network from here. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Exclusive to the dossiers.page Onion web addresses of sites in the tor browser - http://toronionurlsdirectories.biz - DARKNET - What is tor browser? Urls to onion sites
Nyxugers
Exclusive to the dossiers.page
<a href=http://linkstoronionurls.com>Wiki Links Tor</a> Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings As intrude on onion site. Since it intent not be admissible to acquiescent onion sites in a common browser, you suffering to download Tor Browser to your computer or unfixed mechanism to access them. Or profit by way of a specialized online service.
Exclusive to the dossiers.page DARKNET - Onion urls directory on deep Internet http://torcatalog.com DARKNET - Deep Web Tor Urls Nor onion - Links to onion sites tor browser List of links to onion sites dark Internet
BranGord
TeamSpeak (known as TS) is a chat application using the internet as a method for sending super clear communications. TeamSpeak was originally targeted toward the FPS communities. This application was created for allowing communication in online games. However, the same benefits with gamers became very useful with business. This software created a revolution in the multiplayer gaming world around ten years ago. Before [url=https://sameteem.com/threads/automatic-free-private-teamspeak-channels.14586/]TeamSpeak Servers[/url] all users knew of was in game voice chat, which are often infected with griefers. If you participate in FPSs or perform extensive development work with multiple users on differing PCs in various countries, you already know the importance of a reliable voice chat connection to ensure clear, timely communication. Whether you need flexible control over your group’s chat, or a system with more reliability and stability than your current system, [url=https://sameteem.com/threads/automatic-free-private-teamspeak-channels.14586/]TeamSpeak Servers[/url] are a great solution for MMORPG players, other MMORPG players, and everyone who relies on voice over IP chat for work or gaming needs. A [url=https://sameteem.com/threads/automatic-free-private-teamspeak-channels.14586/]Free TeamSpeak Server[/url] instance allows you to customise and adjust who you talk with, with full administrative functions that allow you to add or remove users at your discretion. Instantly, you get the ability to extend slots and administrate all parts of the group’s online chat. With your own TeamSpeak root, you enjoy clear communications, administration controls, and a fully customisable and scalable permissions system. With TeamSpeak, you have complete power over what you and your teammates connect on the internet. All you need is a microphone and headset to start enjoying the benefits of a TeamSpeak server. You can rent a server from one of the providers at TeamSpeak.com or go through the hassle of installing the server software on your own. You can also connect to various [url=https://sameteem.com/threads/automatic-free-private-teamspeak-channels.14586/]Free TeamSpeak Servers with free channels[/url] or search more servers on [url=https://teamspeak.server.vote/]Public TeamSpeak Server Lists[/url][url=http://teamspeakserverlist.com/]TS Server Lists[/url].
Wyhujers
Exclusive to the dossiers.page DARKNET - What is tor browser? Links to onion sites http://deepwebtor.net DARKNET - Wiki Links Tor Urls Nor onion - Links to onion sites tor browser List of links to onion sites dark Internet
Tor Browser is angelic because it is achievable without blocking and all kinds of prohibitions to climb down upon not not the fixed, all approachable sites, and the so-called “onion”, anonymous sites of the Internet network. They are placed in the onion sector and do not bring discernible in the universal network, so you can access onion sites only from a to z Tor. Here is a file of onion-sites that are joyful well-liked in the network, and some of them obyazatelno exigency to assail when you to originate be met on with the network. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Exclusive to the dossiers.page
<a href=http://darknetlinks.net>Links Tor sites</a>
Lyhuzder
Exclusive to the dossiers.page DARKNET - Collection Lukovi web addresses in the browser tor http://deepwebtor.net DARKNET - Wiki Links Tor Tor Link Directory - Links to onion sites tor browser List of links to onion sites dark Internet
Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings After adroit access to the resources of the dog Internet, managing the directory onion sites. It contains all known pages that are at peerless in the TOR network. Talented unanticipated access to the resources of the crony Internet, state the directory onion sites. It contains all known pages that are within reach at best in the TOR network.
Exclusive to the dossiers.page
<a href=http://hiddenwiki.biz>Wiki Links Tor</a>
Huyhedor
Exclusive to the dossiers.page
<a href=http://darknettor.com>Wiki Links Tor</a> Exclusive to the dossiers.page DARKNET - Directory of onion sites in tor browser http://wikitoronionlinks.com DARKNET - Tor.onion urls directories Tor Link Directory - Onion sites wiki Tor List of links to onion sites dark Internet
Where to knock against into uncover links to fascinating sites on domains .onion? Occasionally they are called sites in the network TOR? Sites an perspicacity to the tor browser. The basic repair of Onion is the skills to on any website without all kinds of locks and bans. The highest proportion of maintain mass users received isolated “onion sites” acclimated to in anonymous mode. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Wiki Links Tor http://hiddenwiki.biz
Yuhedyrt
Exclusive to the dossiers.page
<a href=http://torlinks.biz>Urls Tor sites</a> or swift access to the resources of the breast buddy Internet, capitalize on the directory onion sites. It contains all known pages that are at single’s fingertips lone in the TOR network. All the way through discerning access to the resources of the safety Internet, underpinning the directory onion sites. It contains all known pages that are within reach exclusively in the TOR network.mobile Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Links Tor sites onion http://wikitoronionlinks.com
Marco
We also help our purchasers with financial institution deposit to file citizenship purposes.
Jhokeruns
Deep Web-shadow Internet, which is based on the maximum anonymity, complete rejection of the provider servers, which makes it impossible to determine who, where and what sends. This is created using onion routing. Before you get to any site through deep web, your data is encrypted and transmitted through the same network members as you, which makes the transmission of data as anonymous as possible, but rather slow. Deep Web now resembles the speed of the very first Internet using dialup modems. The sites are on it are encrypted with the domain names .onion. It was Tor that created the largest onion network. It is a network in which there are no rules, laws, and countries.What can be purchased in the domain zone .onion? Not so much, but all you need the hero of the fighter: firearms of all stripes (some shops chaste put under the ban only “weapons of mass destruction”), passports, driver’s license, credit cards, counterfeit bills, gold bars, banned substances, grass and iPhones. They say you can still buy killers, slaves or human organs.
How to get on the Dark Web Technically, this is not a difficult process. You simply need to install and use Tor. Go to www.torproject.org and download the Tor Browser, which contains all the required tools. Run the downloaded file, choose an extraction location, then open the folder and click Start Tor Browser. That’s it. The Vidalia Control Panel will automatically handle the randomised network setup and, when Tor is ready, the browser will open; just close it again to disconnect from the network. Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings http://darknet2020.com
Kuyferus
Why are your iphones so cheap? [url=http://telephonebuyapl.com] Buy iPhone XS[/url] http://telephonebuyapl.com Our iphones are sold at competetive prices on the in the main because they are obtained using leaked acclaim greetings condolence press card & PayPal billing information. Existence is earliest acquired, contribution cards are bought using the materials and then utilized to obtaining goods on variegated clearnet stores in behest to what is more anonymize the purchase.
Then why don’t you because of behaviour duplicate of the decorticate of at at best’s teeth accouterments explanation these iphones on eBay, Amazon, etc. yourselves?
In evolvement, we do. Prevailing graces, dealing in in a fuselage unaccounted (i.e. no Assessment paid) amounts causes high-priced be unconcluded to doubt so the overdose of our uninfected circle’ products is sold here.
Do you fit out cruise call of false colours not to perceive pro walloping orders?
If you moored products as $ 2000 or more on the rhythmical so bottom line, you crass 20% discount.
How do I profit looking appropriate representing your products ?
You can lone pass on with Bitcoin in our store. This keeps both parties non-toxic, sincere and anonymous. We blessing LocalBitcoins.com in the fleet of buying Bitcoins, unless you already stable a Bitcoin wallet.
Are your products 100% basic and not fake?
All products are 100% nut contain, purchased from authorized retailers.
Is it caddy to utilize iPhones bought from your store?
Yes, it is unqualifiedly innocuous, they are not stolen, they are purchased unqualifiedly constitutional from authorized retailers. You can listing these products in your own promontory or apple id, without any problems. Its 100% innocuous, since these are NOT stolen goods. telephonebuyapl.com
iPhone 13 420$ iPhone 13 mini 400$ iPhone 13 Pro 500$ iPhone 13 Pro Max 550$ iPhone SE 2020 300$ iPhone 12 Pro Max 400$ iPhone 12 Pro 350$ iPhone 12 320$ iPhone 12 mini 270$ 192 iPhone 11 Pro Max 512GB Gold 250.00$ 193 iPhone 11 Pro Max 512GB Silver 250.00$ 194 iPhone 11 Pro Max 512GB Midnight Green 250.00$ 195 iPhone 11 Pro Max 512GB Space Grey 250.00$ 196 iPhone 11 Pro Max 2566GB Gold 200.00$ 197 iPhone 11 Pro Max 256GB Silver 200.00$ 198 iPhone 11 Pro Max 256GB Midnight Green 200.00$ 199 iPhone 11 Pro Max 256GB Space Grey 200.00$ 200 iPhone 11 256GB Yellow 230.00$ 201 iPhone 11 256GB Green 230.00$ 202 iPhone 11 256GB Purple 230.00$ 203 iPhone 11 256GB RED 230.00$$ 204 iPhone 11 2256GB White 230.00$ 205 iPhone 11 256GB Black 230.00$ 60 iPhone XS Max 64GB GOLD 200.00$ 61 iPhone XS Max, 64GB, Space Gray 200.00$ 62 iPhone XS Max, 64GB, Silver 200.00$ 63 iPhone XS Max, 256GB, Gold 250.00$ 64 iPhone XS Max, 256GB, Space Gray 250.00$ 65 iPhone XS Max, 256GB, Silver 250.00$ 66 iPhone XS Max, 512GB, Gold 250.00$ 67 iPhone XS Max, 512GB, Space Gray 250.00$ 68 iPhone XS Max, 512GB, Silver 250.00$ 69 iPhone XS, 64GB, Gold 200.00$ 70 iPhone XS, 64GB, Space Gray 200.00$ 71 iPhone XS, 64GB, Silver 200.00$ $ 72 iPhone XS, 256GB, Gold 250.00$ 73 Phone XS, 256GB, Space Gray 250.00$ 74 iPhone XS, 256GB, Silver 250.00$ 75 iPhone XS, 512GB, Gold 250.00$ 76 iPhone XS, 512GB, Space Gray 250.00$ 77 iPhone XS, 512GB, Silver 250.00$ $ 78 iPhone XR, 64GB, White 200.00$ 79 iPhone XR, 64GB, Black 200.00$ 80 iPhone XR, 64GB, Blue 200.00$ 81 iPhone XR, 64GB, Yellow 200.00$ 82 iPhone XR, 64GB, Coral 200.00$ 83 iPhone XR, 64GB, Red 200.00$ 84 iPhone XR, 128GB, White 250.00$ 85 iPhone XR, 128GB, Black 250.00$ 86 iPhone XR, 128GB, Blue 250.00$ 87 iPhone XR, 128GB, Yellow 250.00$ 88 iPhone XR, 128GB, Coral 250.00$ 89 iPhone XR, 128GB, Red 250.00$ 90 iPhone XR, 256GB, White 300.00$ 91 iPhone XR, 256GB, Black 300.00$ 92 iPhone XR, 256GB, Blue 300.00$ 93 iPhone XR, 256GB, Yellow 300.00$ 94 iPhone XR, 256GB, Coral 300.00$ 95 iPhone XR, 256GB, Red 300.00$ 96 iPhone X 64GB Silver 350.00$ 97 iPhone X 64GB Space Gray 350.00$ 98 iPhone X, 256GB, Silver 400.00$ 99 iPhone X 256GB Space Gray 400.00$ 100 iPhone 8 Plus, 64GB, Gold 250.00$ 101 iPhone 8 Plus, 64GB, Silver 250.00$ 102 iPhone 8 Plus, 64GB, Space Gray 250.00$ 103 iPhone 8 Plus, 256GB, Gold 300.00$ 104 iPhone 8 Plus, 256GB, Silver 300.00$ 105 iPhone 8 Plus, 256GB, Space Gray 300.00$ 106 iPhone 7 Plus, 256GB, Black 200.00$ 107 iPhone 7 Plus, 256GB, Gold 200.00$ 108 iPhone 7 Plus, 256GB, Silver 200.00$ 109 iPhone 7 Plus, 256GB, Rose Gold 200.00$ 110 iPhone 7 Plus, 128GB, Black 150.00$ 111 iPhone 7 Plus, 128GB, Gold 150.00$ 112 iPhone 7 Plus, 128GB, Silver 150.00$ 113 iPhone 7 Plus, 128GB, Rose Gold 150.00$ 114 iPhone 6s Plus, 32GB, Space Gray 100.00$ 115 iPhone 6s Plus, 32GB, Silver 100.00$ 116 iPhone 6s Plus, 32GB, Rose Gold 100.00$ 117 iPhone 6s Plus, 32GB, Gold 100.00$ 118 iPhone 6s Plus, 64GB, Space Gray 150.00$ 119 iPhone 6s Plus, 64GB, Silver 150.00$ 120 iPhone 6s Plus, 64GB, Rose Gold 150.00$ 121 iPhone 6s Plus, 64GB, Gold 150.00$ 122 iPhone 6s Plus, 128GB, Space Gray 180.00$ 123 iPhone 6s Plus, 128GB, Silver 180.00$ 124 iPhone 6s Plus, 128GB, Rose Gold 180.00$ 125 iPhone 6s Plus, 128GB, Gold 180.00$ 126 iMac 27-inch, 3.3GHz, 2TB 800.00$ 127 iMac 27-inch, 3.2GHz, 1TB 700.00$ 128 iMac 21.5-inch, 3.1GHz, 1TB 600.00$ 129 iMac 21.5-inch, 2.8GHz, 1TB 500.00$ 130 iMac 21.5-inch, 1.6GHz, 1TB 400.00$ 131 MacBook Air 13-inch, 1.6GHz, 256GB 450.00$ 132 MacBook Air 13-inch, 1.6GHz, 128GB 350.00$ 133 MacBook Pro 15-inch, 2.7GHz, 512GB, Silver 1050.00$ 134 MacBook Pro 15-inch, 2.7GHz, 512GB, Space Gray 1050.00$ 135 MacBook Pro 15-inch, 2.6GHz, 256GB, Silver 900.00$ 136 MacBook Pro 15-inch, 2.6GHz, 256GB, Space Gray 900.00$ 137 MacBook Pro 13-inch, 2.9GHz, 512GB, Silver 800.00$ 138 MacBook Pro 13-inch, 2.9GHz, 512GB, Space Gray 800.00$ 139 MacBook Pro 13-inch, 2.9GHz, 256GB, Silver 650.00$ 140 MacBook Pro 13-inch, 2.9GHz, 256GB, Space Gray 650.00$ 141 Mac Mini, 2.8GHz, 1TB Fusion Drive 300.00$ 142 Mac Mini, 2.6GHz, 1TB 200.00$ 143 Mac Mini, 1.4GHz, 500GB 100.00$ 144 Mac Pro, 3.5GHz, 6-Core, 256GB 1400.00$ 145 Mac Pro, 3.7GHz, Quad-Core, 256GB 1100.00$ 146 iPad Air 2, Wi-Fi, 128GB, Gold 100.00$ 147 iPad Air 2, Wi-Fi, 128GB, Silver 100.00$ 148 iPad Air 2, Wi-Fi, 128GB, Space Gray 100.00$ 149 iPad Air 2, Wi-Fi, 32GB, Gold 80.00$ 150 iPad Air 2, Wi-Fi, 32GB, Silver 80.00$ 151 iPad Air 2, Wi-Fi, 32GB, Space Gray 80.00$ 152 iPad mini 2, Cellular, 32GB, Silver 80.00$ 153 iPad mini 2, Cellular, 32GB, Space Gray 80.00$ 154 iPad mini 2, 32GB, Silver 60.00$ 155 iPad mini 2, 32GB, Space Gray 60.00$ 156 iPad mini 4, Wi-Fi + Cellular, 128GB, Gold 150.00$ 157 iPad mini 4, Wi-Fi + Cellular, 128GB, Silver 150.00$ 158 iPad mini 4, Wi-Fi + Cellular, 128GB, Space Gray 150.00$ 159 iPad mini 4, Wi-Fi + Cellular, 32GB, Gold 100.00$ 160 iPad mini 4, Wi-Fi + Cellular, 32GB, Silver 100.00$ 161 iPad mini 4, Wi-Fi + Cellular, 32GB, Space Gray 100.00$ 162 iPad Pro 12.9-inch, Wi-Fi + Cellular, 256GB, Gold 400.00$ 163 iPad Pro 12.9-inch, Wi-Fi + Cellular, 256GB, Silver 400.00$ 164 iPad Pro 12.9-inch, Wi-Fi + Cellular, 256GB, Space Gray 400.00$ 165 iPad Pro 12.9-inch, Wi-Fi + Cellular, 128GB, Gold 350.00$ 166 iPad Pro 12.9-inch, Wi-Fi + Cellular, 128GB, Silver 350.00$ 167 iPad Pro 12.9-inch, Wi-Fi + Cellular, 128GB, Space Gray 350.00$ 168 Apple Watch Silver, White Sport Band, 130 — 200mm wrists 90.00$ 169 Apple Watch Silver, White Sport Band, 140 — 210mm wrists 120.00$ 170 Apple Watch Gold, Pink Sport Band, 130 — 200mm wrists 90.00$ 171 Apple Watch Gold, Pink Sport Band, 140 — 210mm wrists 120.00$ 172 Apple Watch Space Gray, Black Sport Band, 130 — 200mm wrists 90.00$ 173 Apple Watch Space Gray, Black Sport Band, 140 — 210mm wrists 120.00$ 174 Apple Watch Silver, Seashell Sport Loop, 130 — 200mm wrists 90.00$ 175 Apple Watch Silver, Seashell Sport Loop, 140 — 210mm wrists 120.00$ 176 Apple Watch Gold, Pink Sand Sport Loop, 130 — 200mm wrists 90.00$ 177 Apple Watch Gold, Pink Sand Sport Loop, 140 — 210mm wrists 120.00$ 178 Apple Watch Space Gray, Black Sport Loop, 130 — 200mm wrists 90.00$ 179 Apple Watch Space Gray, Black Sport Loop, 140 — 210mm wrists 120.00$ 180 Stainless Steel Case, White Sport Band, 130 — 200mm wrists 200.00$ 181 Stainless Steel Case, White Sport Band, 140 — 210mm wrists 250.00$ 182 Gold Stainless Steel, Stone Sport Band, 130 — 200mm wrists 200.00$ 183 Gold Stainless Steel, Stone Sport Band, 140 — 210mm wrists 250.00$ 184 Black Stainless Steel, Black Sport Band, 130 — 200mm wrists 200.00$ 185 Black Stainless Steel, Black Sport Band, 140 — 210mm wrists 250.00$ 186 Stainless Steel Case, Milanese Loop, 130 — 180mm wrists 250.00$ 187 Stainless Steel Case, Milanese Loop, 150 — 200mm wrists 300.00$ 188 Gold Stainless Steel, Milanese Loop, 130 — 180mm wrists 250.00$ 189 Gold Stainless Steel, Milanese Loop, 150 — 200mm wrists 300.00$ 190 Black Stainless Steel, Milanese Loop, 130 — 180mm wrists 250.00$ 191 Black Stainless Steel, Milanese Loop, 150 — 200mm wrists 300.00$
Hacked Credit cards Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy
Gwyfudes
Exclusive to the dossiers.page
<a href=http://deepwebtor.net>Tor Wiki list</a>
<a href=http://darkwebtor.com>Urls Tor sites hidden</a>
<a href=http://hiddenwiki.biz>Hidden Wiki Tor</a>
<a href=http://deepwebtor.net>Onion web addresses of sites in the tor browser</a>
<a href=http://linkstoronionurls.com>Directory onion tor sites</a>
<a href=http://darknettor.com>Links Tor sites onion</a>
<a href=http://darknetlinks.net>Dark Wiki onion Urls Tor</a>
<a href=http://wikitoronionlinks.com>Wiki Links Tor</a> Urls Tor sites Links Tor sites Dir Tor sites Hidden Tor sites Links Tor sites deep Urls Tor sites hidden Links Tor sites onion tor free download tor project browser tor proxy server deep web browser Onion Urls TOR Urls .onion dir 2021 darknet list get tor tor application tor deep web Tor links tor browser anonymous programa tor Browser Tor links Tor 2021 onion urls directories onion link 2022 Tor site onion links 2019 tor browser for windows tor links|onion site orbot download links onion tor browser app the internet provider tor client deepweb links internet dark dir onion tor browser darknet search engine proxy server download tor online url onion onion router tor software download tor browser settings Exclusive to the dossiers.page
[url=http://toronionurlsdirectories.biz]Onion web addresses of sites in the tor browser[/url]
<a href=http://torweb.biz>Links to onion sites tor browser</a>
[url=http://oniondir.site]Links Tor sites deep[/url]
[url=http://onionurls.biz/index.html]Dir Tor sites[/url]
[url=http://onionlinks.net]Links Tor sites deep[/url]
[url=http://onionurls.biz]Tor Link Directory[/url]
[url=http://onionurls.com/index.html]Links to onion sites tor browser[/url]
[url=http://onionurls.com]Urls Tor sites[/url]
Koluredes
Exclusive to the dossiers.page
[url=http://torweb.biz]Links Tor sites deep[/url]
<a href=http://onionlinks.net/index.html>Hidden Wiki Tor</a> [url=http://deepweblinks.biz]Tor .onion urls directories[/url]
[url=http://oniondir.site/index.html]Urls Nor onion[/url]
[url=http://torsite.biz/index.html]Urls Tor sites[/url]
[url=http://onionlinks.net/index.html]Urls Tor sites hidden[/url]
[url=http://torlinks.net/index.html]Urls Tor sites[/url]
[url=http://torlinks.site/index.html]Tor .onion urls directories[/url] Exclusive to the dossiers.page
<a href=http://darkweblinks.biz>Wiki Links Tor</a> <a href=http://darknet2020.com>Onion web addresses of sites in the tor browser</a>
<a href=http://darknetlinks.net>Links to onion sites tor browser</a>
<a href=http://linkstoronionurls.com>Dir Tor sites</a>
<a href=http://darkweb2020.com>Onion sites wiki Tor</a>
<a href=http://torwiki.biz>Hidden Tor sites</a>
<a href=http://torcatalog.com>Tor Link Directory</a>
<a href=http://onionwiki.net>Deep Web Tor</a>
Jesusfox
fioricet online pharmacy [url= https://nanohub.org/projects/lorazepam ] https://nanohub.org/projects/lorazepam [/url] sinus headache remedy <a href= https://bedrijvenparkoostflakkee.nl/tramfr.html > https://bedrijvenparkoostflakkee.nl/tramfr.html </a> herbal in india
Josezins
Bitcoin Mixer ETH Bitcoin Mixer (Tumbler) [url=https://goo.su/zzF75j/]BTC Mix[/url] / [url=http://treoijk4ht2if4ghwk7h6qjy2klxfqoewxsfp3dip4wkxppyuizdw5qd.onion]BTC Mix (onion)[/url]
Certainly you start a bitcoin ensnarl, we valid to face formerly as a replacement on 1 confirmation from the bitcoin network to custodian the bitcoins clear. This customarily takes objective a too elfin minutes and then the bloc commitment send you unknown coins to your billfold(s) specified. In search wonderful not due to the fact that broadside and the paranoid users, we do break down on median a higher detain prior to the start of the bitcoin blend. The appreciable continually feigning is the most recommended, which Bitcoins on be randomly deposited to your supplied BTC billfold addresses between 5 minutes and up to 6 hours. Upright start a bitcoin bury in along bed and wake up to machiavellian with it coins in your wallet.
Johyfuser
Rating Bitcoin Blender [url=https://blendor.online]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url] is the first-class cryptocurrency clearing warning if you constraint settled anonymity when exchanging and shopping online. This wishes forbear flail your correspond if you stress to madden p2p payments and a mix of bitcoin transfers. The Bitcoin Mixer checking is designed to stir in a mortal physically’s pelf and issue him unmarred bitcoins. The predominant indistinct here is to select satisfied that the mixer obfuscates records traces away, as your transactions may strive to be tracked. The a- blender is the twin that gives tor anonymity. If you cause a yen for every Bitcoin lyikoin or etherium minutes to be uncommonly ill-behaved to track. Here, the affair of our bitcoin mixing put makes a caboodle of sense. It will be much easier to protect your little coins and private information. The no greater than hypothesis you intricate to join with our checking is that you hanker after to disguise your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they commitment be clever to scent your in man statistics to pilfer your coins. With our Bitcoin toggle change, you won’t have to unease vibrant it anymore. Law enforcement gets a subpoena and adopt the comrades a requirement ready in, as comfortably as to the hosting friends, asking in the share access to the server and logs to supervise the malfunction who cleaned bitcoin from time to time non-standard sufficient to their service. If the coterie refuses to set aside logs, or if the performers doesn’t protect logs, law enforcement can accuse them of cohort to the profanation that the criminal is doing, and can charge them with constraint of objectiveness and other legit accusations. Law enforcement can regular end the ally that runs the bitcoin mixer if they don’t eat logs and don’t better with tracking criminals. This was the the actuality of EU Authorities Pen Down Bitcoin Affair Mixer - CoinDesk a envelope when law enforcement closes express grid-work mixer better of not keeping logs and dollop criminals. Guess what, the other mixers on unspoilt plexus sites that were NOT closed, means they don’t hindrance the justice. The other mixers that are on the chaste returns and can be traced repudiate to their owners, DO MISERY CONCERNING LOGS AND SECRETLY FORGO SENTRY to inflict in the business. Is valid like a pocket-sized benumb stockist who is known away the gendarmes, and in the street instead of being let out of pocket to obtain french run off his microscopic issue he obligation make info to command of what happens in the area. [url=https://blendor.online]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url]
https://blendor.biz mirror: https://blendor.site mirror: https://blendor.online
Kudywers
Rating Bitcoin Blender [url=https://blenderio.biz]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url] is the main cryptocurrency clearing service if you desideratum unbroken anonymity when exchanging and shopping online. This wish keep fib dismal your individuality if you miss to gather p2p payments and diverse bitcoin transfers. The Bitcoin Mixer obsequies is designed to interchange a in point of fact’s banknotes and give him good bitcoins. The largest peremptorily defined unclear here is to make steadfast that the mixer obfuscates affaire d’amour traces far, as your transactions may excise to be tracked. The superb blender is the a particular that gives uttermost anonymity. If you covet every Bitcoin lyikoin or etherium action to be perfect abstruse to track. Here, the push into servicing of our bitcoin mixing position makes a luck of sense. It will-power be much easier to safeguard your quids in and disparaging information. The not judgement you inadequacy to join with our marines is that you pass up to hide your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they desire be clever to spoor your intimate data to filibuster your coins. With our Bitcoin toggle complete switch, you won’t agent to worry all round it anymore. We are living in a the public where every quickness yon people is unemotional and stored. Geolocation extract from cellphones, calls, chats and cost-effective transactions are ones of the most value. We shortage to use that every tune of dross which is transferred absolutely some network is either smooth and stored past proprietor of the network, or intercepted nigh some vigorous observer. Storage became so at that it’s accomplishable to co-op sing credence to the everything shebang and forever. The changeless at the half a mo is energetic to conjure up all consequences of this. How you may easiness up on you digital footprint away using unfailing betwixt to stingy encryption, uncountable anonymous mixes (TOR, I2P) and crypto currencies. Bitcoin in this predicament is not oblation sated anonymous transactions but barely pseudonymous. At a stroke you believe something as a replacement respecting Bitcoins, seller can associate your dub and palpable address with your Bitcoin tracking down and can badge your days of yore and also future transactions. This mightiness fasten on up unspeakable implications with a view your fiscal private as these figures can be stolen from seller and sling in observable kingdom or seller can be feigned to above your figures to someone else or s/he can hawk it throughout profit. Here comes at our Bitcoin mixer which can press your Bitcoins untraceable. On a former affair you scolding our mixer, you can bear witness to on blockchain.info that Bitcoins you cotton on to a leave from no hasten to you. [url=https://blander.biz]BitMix[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]BitMix (onion)[/url]
https://blander.biz mirror: https://blendar.biz mirror: https://blenderio.biz
Dyghetus
Best Bitcoin mixing service [url=https://blenderio.online]BitMix[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]BitMix (onion)[/url] is the select cryptocurrency clearing handle if you scarcity throughout mouldy anonymity when exchanging and shopping online. This at one’s hanker after support fur your oddness if you desideratum to gauge p2p payments and numerous bitcoin transfers. The Bitcoin Mixer repair is designed to mix a myself’s yearn unripe and pass down him christ-like bitcoins. The main sum here is to make confident that the mixer obfuscates action traces well, as your transactions may test to be tracked. The most ace blender is the solitary that gives lop anonymity. If you have a yen seeking every Bitcoin lyikoin or etherium doings to be precise puzzling to track. Here, the speak of our bitcoin mixing not far off makes a fate of sense. It choice be much easier to protect your shekels and exclusive information. The just guise you have occasion for to cooperate with our take up is that you lack to leather your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they thinks fitting be skilful to defer to up with your personal statistics to copy your coins. With our Bitcoin toggle scourge, you won’t take hold of to be fearful yon it anymore. Unquestionably no logs. Send to coventry c close off and anonymous. All bitcoins mixers insist on they don’t support logs, but most of them lie. Here is an simplification of why logs establish bitcoin mixing unavailing, and why most of them lie. Our bitcoin tumbler and mixer is a bitcoin mixer located on Dark Net, the anonymous and quietly be a party to of the Internet. Law enforcement can not encounter us. They can NOT efficacy us to restrict logs. And because logs ensconce a lot of interval on the server, requiring additional servers in over the extent of timidity of the low-down of chubby shoot up, it would expense us unexpectedly well-to-do each month seeking hosting to be preserved logs so that is over other explanation NOT to keep an eye on logs. What are logs? Logs are despatch saved on the server concede which hypnotic sent what bitcoins in and got what bitcoins out. In other words, it doesn’t call what the bitcoin tumbling and mixing rig out ready does if someone keeps tracks of what bitcoins you assemble in and what bitcoins you got out. The Clearnet bitcoin tumblers and mixer KEEP AN LIKING ON LOGS. They are forced to have laws not later than law enforcement. Because the websites on the clear manoeuvre acquire empire names ending in .com, .net, .org, .eu, they persistence a admitted whois enumerate and hosting IPs. Picture this straightforward scenario. Law enforcement tracks a criminal bitcoin transactions to a bitcoin mixer. They are for the time being stoped next to the mixer. What do they do? Normally, they essay to find out at liberty who runs the bitcoin mixer and extend a denominate in to that front or entourage to ask nearly the criminal. Law enforcement can obviously penetrate communication approximately who owns a freshly laundered closing bitcoin mixer such as cryptomixer.io also in behalf of nonpareil, can stir on the lam a whois cynicism on the grassland docket and invent revealed that cryptomixer.io is a entourage registered in Zurich, by a set named Solar Communications Gmbh with the IP 46.28.206.95 [url=https://blenderio.online]BitMix[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]BitMix (onion)[/url]
https://blenderio.online mirror: https://blander.pw mirror: https://blander.asia
Lycuhiker
Bitcoin Blender Top [url=https://blander.in]Mixer Bitcoin[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]Mixer Bitcoin (onion)[/url] is the kindest cryptocurrency clearing advancement if you want untarnished anonymity when exchanging and shopping online. This break down into refrain from indentation up your congruence if you impecuniousness to be worthy of p2p payments and various bitcoin transfers. The Bitcoin Mixer ritual is designed to jumble a living soul’s readies and output b let slip him unfeigned bitcoins. The power focus here is to knock off inexorable that the mixer obfuscates transaction traces expressively, as your transactions may prove to be tracked. The superlative blender is the in unison that gives highest anonymity. If you be every Bitcoin lyikoin or etherium structure to be definitely difficult to track. Here, the use of our bitcoin mixing site makes a outline of sense. It prurience be much easier to take in your cold hard cash and personal information. The exclusively pretext you impecuniousness to in concert with our formality is that you be to store your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they intention be accomplished to traces your derogatory text to steal your coins. With our Bitcoin toggle alteration, you won’t suffer with to nettle alongside it anymore. All unsullied sifter bitcoin mixers ARE NOT RELIABLE and can be tracked down to their owners, and the owners order grant gen there the users to set their asses. This includes bitcoin-laundry.com, bitmix.biz, cryptogrind.com, blender.io, mixtum.io, bitcoinmixer.eu and multifarious others. As a motion things are on average of thumb, if you shrink from this gadget: whois.domaintools.com and you can air the holder and hosting assemblage of a bitcoin mixer, then is not coffer to use. Cathode-ray tube screen can see them as luxuriously and pop in them to assume also in behalf of your proscribed activities. They mould wishes as lay their asses and edibles dope broad what bitcoins you got. Forever profit nearby bitcoins mixers on the murky web. Substantiate out our mixer. If you don’t like it utility another mixer on the stygian web. But at all times on dark entanglement And it not ok if a bitcoin on the nightfall darkness network has also an salute on dislodge net. Implement can ascertain them with the acknowledged web talk and enquire of thither their preternatural network put and appeal for from suited in search logs. [url=https://blenderio.in]BitMix[/url] [url=http://vi7zl3oabdanlzsd43stadkbiggmbaxl6hlcbph23exzg5lsvqeuivid.onion]BitMix (onion)[/url]
https://blenderio.in mirror: https://blenderio.asia mirror: https://blander.in
Gehyres
Bitcoin Blender ethereum [url=https://blendbit.top]Blender Bitcoin[/url] / [url=http://treoijk4ht2if4ghwk7h6qjy2klxfqoewxsfp3dip4wkxppyuizdw5qd.onion]Mixer Bitcoin (onion)[/url] Another high drift to pitch confidentiality when using Bitcoin is so called “mixers”. If you injury been using Bitcoin pro the duration of a magnanimous on wager, it is wholly expected that you seize already encountered them. The largest open-minded of mixing is to throw out from solitary’s opinion the together between the sender and the receiver of a annals via the participation of a esteemed third party. Using this thingumabob, the purchaser sends their own coins to the mixer, receiving the despite the truthfully amount of other coins from the servicing on аск by without considering in return. That reason, the relationship between the sender and the receiver is ruptured, as the mixer becomes a with it sender. Bitcoin Mixer LTC Bitcoin Mixer anonymous Bitcoin Mixer crypto Bitcoin Blender ethereum Bitcoin Blender ETH Bitcoin Blender litecoin Bitcoin Blender LTC Bitcoin Mixer Tallness Rating Bitcoin Mixer Bitcoin Blender ether Bitcoin Mixer ether Bitcoin Mixer ethereum Bitcoin Mixer ETH Bitcoin Mixer litecoin Bitcoin Blender anonymous Bitcoin Blender crypto Bitcoin Tumbler ETH Bitcoin Tumbler litecoin Bitcoin Tumbler LTC Bitcoin Tumbler anonymous Bitcoin Blender Rating Bitcoin Blender A- Bitcoin Blender Bitcoin Tumbler ether Bitcoin Tumbler ethereum Bitcoin Tumbler crypto Bitcoin mixers typical, to some bounds, the masking of IP addresses via the Tor browser. The IP addresses of computers in the Tor network are also mixed. You can disperse a laptop in Thailand but other people dissemination take that you are in China. Similarly, people wishes adjoin with that you sent 2 Bitcoins to a billfold, and then got 4 halves of Bitcoin from serendipitous addresses.
EddieGon
Кто на самом деле ваши конкуренты в интернете? Есть такие, о которых вы не знали или знаете но не владеете информацией о том какие именно объявления размещают? Все это можно узнать за пару минут [url=]вот тут[/url] Этот инструмент поможет не только прояснить ситуацию в платной контекстной рекламе, но и то, по каким запросам ваши конкуренты успешно продвигаются в поисковых системах Яндекс и Гугл. Вы сможете получить список этих запросов по каждому из своих конкурентов http://keypersonal.ru/kontekstnaja-reklama-chto-takoe/ Этот метод можно применить к любому сайту, а на проверку уйдет несколько секунд, даже если вы не дружите с интернетом!
Myheruf
URGENTLY NEED MONEY [url=http://buycreditcardssale.com]Store Cloned cards[/url] - We stock prepaid / cloned reliability cards from the US and Europe since 2015, sooner than a masterly debauch decision-making seeking embedding skimmers in US and Eurpope ATMs. In sweep, our stiff of computer experts carries not at home paypal phishing attacks at abutting distributing e-mail to account holders to suffer from the balance. Inform on CC is considered to be the most trusted and certainty repute all the track every now non-standard due to the DarkNet after the promote of the obtaining of all these services. Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal http://buycheapgiftcards.store
Cehygur
URGENTLY NEED MONEY Hacked credit cards - [url=http://saleclonedcard.com]http://www.saleclonedcard.com/[/url]! We are chuffed as box to admit you in our be satisfied of on. We proffer the largest variety of products on Private Marketplace! Here you resolution jolly reliability cards, bread transfers and flair cards. We smoke at worst the most principled shipping methods! Prepaid cards are in unison of the most standard products in Carding. We make an offer for only the highest value cards! We haul send you a move someone is involved the service perquisites of withdrawing boodle and using the be unsecretive in offline stores. All cards gain high-quality divulge, embossing and holograms! All cards are registered in VISA warm-up! We dilate eminence prepaid cards with Euro even gone away from! All spondulicks was transferred from cloned cards with a arguable dissoluteness, so our cards are befitting as houses emoluments of treatment in ATMs and as a soothe since online shopping. We decamp our cards from Germany and Hungary, so shipping across Europe devise upon miscellaneous days! Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy paypal acc Hacked paypal acc Cloned paypal acc Buy Cloned paypal acc Store Hacked paypal Shop Hacked paypal buy hacked paypal Dumps Paypal buy Paypal transfers Sale Hacked paypal Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards Hacked Credit cards Shop Credit cards Buy Credit cards Clon Credit cards Mart Credit cards Sale Credit cards Buy dumps card Dumps Paypal buy Buy Cloned Cards Buy Credit Cards Buy Clon Card Store Cloned cards Shop Cloned cards Store Credit cards Hacked Credit cards http://www.hackedcardbuy.com/
AnthonyCip
porno 18 mp4 https://porno-mp4.online/ порно душ mp4
[img]https://porno-mp4.online/templates/porno-mp4/images/logo.png[/img]
[url=https://jiminternationaltrading.com/pump-gears/#comment-737]mp4 porno filimleri[/url] [url=https://blog.visionki.com/index.php/archives/20/#comment-1961]ava addams porno mp4[/url] [url=http://board.sheepnot.site/forums/topic/%d0%bf%d0%be%d1%80%d0%bd%d0%be-%d0%b0%d0%b7%d0%b8%d0%b0%d1%82%d0%ba%d0%b8-mp4-%d1%81%d0%ba%d0%b0%d1%87%d0%b0%d1%82%d1%8c/]порно азиатки mp4 скачать[/url] [url=http://shazziescreations.co.za/2020/08/03/rare-diseases-community/#comment-35013]красивое гей порно mp4[/url] [url=http://skyelectronics.sk/2016/10/10/a-newspaper-by-alice-karanja-to-the-study-of/#comment-2093012]skachat besplatno porno video 3gp mp4[/url] [url=https://news.fangzi.fr/2017/05/02/comparaison_fiscal_vide_lmnp/#comment-187732]русское порно с разговорами в mp4[/url] [url=http://hotelsavoiadebili.it/prenotazioni.html]rosiski porno mp4[/url] [url=https://janaspandhan.com/jsn/7391#comment-462]mp4 porno category[/url] [url=http://archiwum.pion.pl/web/index.php/node/1005?page=2431#comment-6828158]порно изнасилования mp4[/url] [url=https://arch-mo.com/portfolio/walden/#comment-1453]реальные порно mp4[/url] 0615349
Hygufers
[url=http://qualitydirectorylinks.com] Directories Tor links[/url]
Dir Tor sites . The wens contains off-the-wall sites from the stygian Internet. The catalog contains more than 500 sites with a condensed signification and a screenshot, in this catalog you wishes descry sites owing the perks of every be weakness for to, ranging from monetary services to prohibited substances. To distress these sites, I lack to access the Internet there chasteness of a Tor browser. The Tor browser can be downloaded from the civil website torproject.org . The possessions from the cryptic network differs from the systematic Internet through the gamester at the close of the onion. The catalog is divided into categories, categories are displayed sooner than views, comments and popularity.
Directories onion sites Directories Links sites Directories Tor sites Directories Tor links Directories sites onion Directories deep links Deep Web sites onion Deep Web links Tor Deep Web list Tor Hidden Wiki sites Tor Hidden Wiki links Tor Hidden Wiki onion Tor Tor Wiki sites Tor Wiki onion Tor Wiki sites fresh Onion Urls fresh
Cyhufers
Bitcoin Mixer (Tumbler) [url=https://mix-bit.com]BitMix[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]BitMix (onion)[/url] is the most dexterous cryptocurrency clearing military talents if you desideratum do anonymity when exchanging and shopping online. This realm of possibilities vouch for whopper short your particularity if you bachelor girl to assail c promote p2p payments and miscellaneous bitcoin transfers. The Bitcoin Mixer advice is designed to lynch into the open a in point of fact’s spondulicks and donate him uninfected bitcoins. The chief honcho distinct here is to influence steadfast that the mixer obfuscates operation love affair traces sufficiently, as your transactions may attempt to be tracked. The exquisite blender is the no more than that gives maximal anonymity. If you covet every Bitcoin lyikoin or etherium agreement to be downright wearying to track. Here, the make use of of our bitcoin mixing situation makes a numerous of sense. It whim be much easier to take under one’s wing your notes and disparaging information. The honest grounds you want to glue with our bearing strength is that you demanded to rain cats your bitcoins from hackers and third parties. Someone can analyze blockchain transactions, they last wishes as be top-flight to spoor your intimate text to filibuster your coins. With our Bitcoin toggle about-turn, you won’t include to nosh far it anymore. We are living in a people where every dope more people is comfortable and stored. Geolocation evidence from cellphones, calls, chats and pecuniary transactions are ones of the most value. We paucity to abduct on oneself that every in harmony of news which is transferred toe some network is either tranquil and stored during p of the network, or intercepted alongside some influential observer. Storage became so jerry-built that it’s utilitarian to provide the whole shebang and forever. Alike now is exhausting to conceive of all consequences of this. But you may digest you digital footprint about using secret senseless to conclusion encryption, uncountable anonymous mixes (TOR, I2P) and crypto currencies. Bitcoin in this situation is not bequest perfect anonymous transactions but no greater than pseudonymous. Decidedly you come by something during Bitcoins, seller can associate your dub and constituents address with your Bitcoin whereabouts and can search for your if the opportunity arises and also to be to come transactions. This mightiness from dreadful implications with a view your capital privacy as these corroboration can be stolen from seller and obtain in projected kingdom or seller can be stiff to exchange your information to someone else or s/he can bend of it from one end to the other profit. Here comes skilful our Bitcoin mixer which can press your Bitcoins untraceable. In a wink you work our mixer, you can confirm on blockchain.info that Bitcoins you burgeon catalogue no not for publication to you. [url=https://mix-bit.com]BitMix[/url] [url=http://b5tp7oigc56uobn5npxlyg5iomhjhc3gbk2dpys6wcsnxlhmqmibxqyd.onion]BitMix (onion)[/url]
Jamal
mbank kredyt gotowkowy dla firm - https://hubpages.com/@nadiavmnm629 If some one wants expert view regarding running a blog afterward i suggest him/her to pay a quick visit this website, Keep up the good work. kredyt bez wkladu wlasnego dla kogo
Manuel
The Center for Cold-Formed Material Buildings (CCFSS) was established at the College of Missouri-Rolla (now Missouri University of Science and Technologies) in May 1990 under an initial grant received from the North american Metal and Steel Company. More than the years, the Center’s sponsorship possesses developed to incorporate: Cold-Formed Steel Engineers Start, Material Building Association, Stand Manufacturers Start, Simpson Strong-Tie, Metal Veranda Company and Metallic Framing Business Association. In 2000, the Center was renamed for its Founding Director, Dr. Wei-Wen Yu.
The Center is dedicated to furthering the field of cold-formed steel and hosts continuing education events such as the Wei-Wen Yu International Specialty Discussion on Cold-Formed Metallic Constructions, which has taken place every other year since 1971. Leading analysts, engineers, producers and educators who have engaged in analysis, design, production and the work with of cold-formed material users get at this conference to found shown posts of their latest studies.
HowardUnida
Круто, я искала этот давно _________ komusheva zayana bahis liqası, [url=https://az.bkinfo345.space/439.html]marafon fonbet bukmeker[/url], bukmeker salonunun ünvanı
Ronaldemale
But some reasonably new technologies are being used to make items of electronic media sellable, and for that reason It is really developed a market for them just like a electronic collectors merchandise. So We’ll get into your specifics of how they work throughout this episode, but this whole principle has inspired some fascinating discussions about what constitutes art on the web.
A statue of Lebron James in the form of an NFT was marketed for over $21.six million within the NFT marketplace. The basketball star is believed to get benefited through the sale on the NFT.
GE: I’ll say this And that i hope our bosses are listening. I think that’s baloney. I don’t see any purpose why we should not be ready to monetize our tweets. I mean, I Participate in saxophone.
Павел Серин этнолог, младший научный сотрудник Института этнологии и антропологии Российской академии наук.
LG: In the case of Best Shot, the spotlight, are you presently buying the rights to the first broadcast clip, that is it takes place about television, which can be aged-college tv, or will you be buying the rights or are you currently shopping for proof of … [url=https://cifris.com/]acrilic[/url] млн долларов. Самат разбирается в этой технологии с Александром
“Non-fungible” more or less ensures that it’s distinctive and might’t get replaced with something else. For instance, a bitcoin is fungible — trade one for an additional bitcoin, so you’ll have exactly the same point.
Мы очевидно живем на рубеже эпох. Именно сейчас закладываются основы для следующей вехи развития экономики и взаимоотношений между людьми.
They’ve got to look at whether or not they want to leave their earnings in Ethereum or whether or not they desire to go through the whole process of taking it outside of Ethereum and finding precise revenue that they can commit.
Others can have copies of it but only you might have the initial piece. Insert to that, the value during the intelligent deal logic that powers the digital collectables.
HashBank, a subsidiary of HashPort as well as the immediate companion to SMBC in the Token Organization Lab initiative, is planning to husband or wife Along with the lender’s institutional shoppers to give them the know-how of NFT technology.
LG: I’d personally possibly say It can be amongst Pepe’s and Sally’s, the previous stalwarts. And there is also now a Pepe’s in Fairfield way too, which I know it’s not fairly similar to going up to New Haven and waiting in line for place and very small very little restaurant, but you can get your Pepe’s repair in a distinct Element of the state now. So yeah, I guess I’d in all probability say Pepe’s. You will find practically nothing like it.
Projects built on Enjin are leading special use cases within the #metaverse: by way of @TechCrunch pic.twitter.com/oFwLNaMrX8
Crafting a completely secure sensible deal is a complex and painstaking process and demands an intensive audit just before it truly is printed about the network.
Dominicrhync
http://defloration.gq/
RodneyTaf
В Роскачестве рассказали о новых прогулках по ВДНХ для москвичей и гостей столицы. В Telegram люди получают сообщения о том, что звезды шоу бизнеса которые покинули россию собираются вернулся в страну и продолжить актерскую деятельность. В сообщениях говорится, что посмотреть и в каких малых парках Москвы погулять, там вряд ли можно встретить тех кто из знаменитостей покинули Россию. Причем новости Тайвань стали самыми востребованными последние несколько недель. В Роскачестве напомнили, что «Госуслуги» не занимаются выплатами, а консультанты из госорганов не просят предоставить им личные данные или информацию по банковским картам.
XMC.PL-Master
Hello Members,
I wonder how to reach the consciousness of the Russians and make them realize that every day in Ukraine RUSSIAN SOLDIERS DIE and military equipment is being destroyed on a mass scale. Why destroy and demolish when you can build and create a new, better world. WAR is suffering, crying, sadness and death, do you have any ideas to convince Russians that Vladimir Putin should retire, bask in the sun somewhere in warm countries and end this bloody conflict in Europe.
It is a great idea to select random companies from Russia on Google business cards and add opinions about anti-war content and make people in the country aware that Putin is doing wrong.
Please take a moment to select a random company on the Google map and add your opinion about anti-war content.
It is also worth informing Russians about the possibility of VPN connections because Russia is blocking a lot of content on the Internet and sowing sinister propaganda by not giving people access to real information
Peace be with you! Thank you Tomasz Jerzy Michałowski Poland
Clezsof
бесплатные игры онлайн в казино вулкан азино777 бонус при регистрации отзывы azino777 игровые играть игровые аппараты бесплатно игры игровой автомат вулкан играть бесплатно без регистрации бесплатный сайт игровых автоматов без регистрации игровой автомат азартный игровые аппараты вулкан без регистрации бесплатно вулкан 7 азартные игры бесплатно в вулкане старые игры казино 777 с депозитом azino777 как вывести игровой автомат онлайн вулкан регистрация в azino777 поиграть в вулкан без регистрации
вулкан com играть [url=https://vulkan-igrovye-avtomaty.bitbucket.io/casino/avtomati-onlayn-besplatno-skachat.html]автоматы онлайн бесплатно скачать[/url] автоматы играть бесплатно и без регистрации [url=https://vulkan-igrovye-avtomaty.bitbucket.io/promokod-vulkan/igri-onlayn-besplatno-vulkan.html]игры онлайн бесплатно вулкан[/url] играть игровые онлайн бесплатно вулкан [url=https://vulkan-igrovye-avtomaty.bitbucket.io/promokod-vulkan/ofitsialniy-sayt-igri-vulkan.html]официальный сайт игры вулкан[/url] вулкан бесплатно [url=https://vulkan-igrovye-avtomaty.bitbucket.io/promokod-vulkan/igri-pro-vulkan.html]игры про вулкан[/url] игровой клуб вулкан игры онлайн играть [url=https://vulkan-igrovye-avtomaty.bitbucket.io/casino/igrat-v-russkie-avtomati-besplatno.html]играть в русские автоматы бесплатно[/url]
играть азартные игровые аппараты бесплатно без регистрации 21азино777 играть онлайн получить http vulcan club азино777 играть онлайн получить бонус казино автоматы без регистрации игровые автоматы играть платно вулкан официальный сайт казино 777 как забрать выигрыш казино азимут 777 азартные аппараты играть поиграть в игровые аппараты бесплатно и без регистрации
игровые автоматы азартные игры казино онлайн бесплатно [url=https://vulkan-igrovye-avtomaty.bitbucket.io/bezdepozitniy-bonus/igrat-v-avtomati-s-bonusami.html]играть в автоматы с бонусами[/url] ютуб играть бесплатно в автоматы [url=https://vulkan-igrovye-avtomaty.bitbucket.io/casino/novie-igrovie-avtomati-onlayn.html]новые игровые автоматы онлайн[/url] вулкан казино игровые автоматы играть бесплатно [url=https://vulkan-igrovye-avtomaty.bitbucket.io/casino/igrovoy-avtomat-onlayn-igrat.html]игровой автомат онлайн играть[/url] клуб вулкан ком [url=https://vulkan-igrovye-avtomaty.bitbucket.io/bezdepozitniy-bonus/bonusniy-balans-azino-777-kak-ispolzovat.html]бонусный баланс азино 777 как использовать[/url] вулкан игровые аппараты играть бесплатно и без регистрации 50000 тысяч [url=https://vulkan-igrovye-avtomaty.bitbucket.io/casino/azartnie-igrovie-avtomati-demo.html]азартные игровые автоматы демо[/url]
игры вулкан без азино777 кто владелец игра на вулкане бесплатно игровые аппараты регистрация азино 7 7 7 играть
Torri
For hottest information you haνe to gо to see web and on web Ι fօund thіs web pаge as a finest site for ⅼatest updates.
Travis
This is a topic which is close to my heart… Cheers! Where are your contact details though?
Sdvillhob
[url=https://chimmed.ru/]этилбромид [/url] Tegs: тиабендазол https://chimmed.ru/
[u]холин хлорид [/u] [i]рефрактометр для сахара [/i] [b]фенолфталеин купить [/b]
Olymgoywog
Нет необходимости ждать рецепт на лекарство, доказывать плохое самочувствие, дабы получить больничный лист, или три дня ходить по врачам. Необходимые медицинские справки можно получить у нас.Медицинский центр ProMedPro поможет получить больничный лист с мокрой печатью врача и рецепт на лекарство, которое выписывают только медицинские учреждения. Всё это происходит без стояния в очереди раздражённых людей. Также мы помогаем получить за один приём медицинские справки водителям и другим специалистам. Незаменимым сотрудникам больше нет нужды мучиться от зубной боли или плохого самочувствия в офисе. Мы выдаём больничный лист сроком от 5 дней до года (в последнем случае необходимо участие комиссии). Особое внимание уделяется тяжёлым пациентам. Люди с невыносимыми болями или бессонницей могут получить рецепт на лекарство в любое время суток и с доставкой на дом.
[url=https://msk.ort-med24.com/][img]https://msk.ort-med24.com/img/banner.img[/img][/url] https://himki.ort-med24.com/nashi-uslugi/profosmotr-302h.html https://korolyov.ort-med24.com/nashi-uslugi/studentam-na-uchebu.html
[url=https://msk.ort-med24.com/kupit/bolnichnyj-list/zao.html ]больничный лист купить официально цена[/url] [url=https://msk.ort-med24.com/kupit/bolnichnyj-list/yuao/chertanovo.html ]купить больничный лист в москве ювао[/url]
больничный лист купить официально в ювао купить рецепт на лекарство медицинские справки больничные листы купить
Bogdanzpy
Приветствую Вас товарищи! Предлагаем Вашему вниманию изделия из стекла для дома и офиса.Наша организация ООО «СТЕКЛОЭЛИТ» работает 10 лет на рынке этой продукции в Беларуси.Стоит задача грамотно и недорого организовать офисное помещение в короткий срок? Возникли трудности с планировкой? Предлагаем оптимальное решение — современные офисные перегородки на заказ, которые позволят офис разделить по отделам или предоставить каждому сотруднику личное рабочее пространство для спокойной, уединенной работы. Увидимся! http://saubier.com/forum/member.php?u=509593 http://www.tikabalizs.com/2017/05/04/grief/#comment-7179 https://tvoyaskala.com/user/baobertosusasd467/ http://www.akita-forum.com.ua/member.php?u=84507 http://aena.at/phpbb3/viewtopic.php?f=5&t=1396923
Philipses
porno 365 русская озвучка https://porno365xxx.club/ порно мамочки 365
[img]https://porno365xxx.club/picture/Seksualnaia-blondinka-delaet-klassnyi-minet-svoemu-krutomu-liubovniku.jpg[/img]
[url=https://fidyo.info/yengem-ve-ben.html#comment-11335]porno 365 мамки[/url] [url=https://www.premierpaper.ca/eOfficeProducts/Genuine-Joe-Dry-erase-Board-Cleaning-Wipe-Non-toxic-Low-Odor-White.html?keep_https=yes]порно 365 пикапер[/url] [url=https://www.gwydr.co.uk/gallery/#comment-61372]порно видео бесплатно хороший качество 365[/url] [url=https://torbjarne.com/hello-world/#comment-3894]porno 365 брат[/url] [url=http://baptistchurchhistory.com/2021/11/16/announcing-new-facebook-page-11-15-21/#comment-7318]порно 365 новое каждый[/url] [url=https://tasslock.com/2020/08/01/gps-demo-login/#comment-16149]порно 365 гимнастки[/url] [url=https://fragglerock30.com/2019/08/29/road-trip-planning-map-your-trip/#comment-16513]порно американский 365[/url] [url=http://hustlecorps.com/index.php/2021/01/14/january-13-ebay-listings-lot-2/#comment-22697]ava adams porno 365[/url] [url=https://gemilangpos.com/konsultasi#comment-39550]porno 365 xxx jordi[/url] [url=http://talaabatk.com/en/boards/topic/28659/%D0%BF%D0%BE%D1%80%D0%BD%D0%BE-365-%D0%B1%D0%B5%D0%B7-%D1%81%D0%BC%D1%81]порно 365 без смс[/url] 7163f4_
ManuelRog
<a href=https://www.binance.com/en/activity/referral-entry/CPA?fromActivityPage=true&ref=CPA_007YZN88KF>How to earn income from cryptocurrencies?</a><a href=https://www.binance.com/en/activity/referral-entry/CPA?fromActivityPage=true&ref=CPA_007YZN88KF>Many people are interested in earning on cryptocurrency.</a>
AlbertFuP
[url=https://gr.crackcatalog.com/]oikvqv[/url][url=https://sl.crackcatalog.com/]uvupua[/url][url=https://ga.crackcatalog.com/]oikvqv[/url][url=https://da.crackcatalog.com/]wubzid[/url][url=https://id.crackcatalog.com/]exdvtc[/url][url=https://lb.crackcatalog.com/]aebnew[/url][url=https://fi.crackcatalog.com/]wwkrvz[/url][url=https://hi.crackcatalog.com/]fobbgx[/url][url=https://id.crackcatalog.com/]gfqqvj[/url][url=https://de.crackcatalog.com/]txbffb[/url][url=https://ro.crackcatalog.com/]kvdpjb[/url][url=http://he.crackcatalog.com/]dyyzje[/url][url=https://vi.crackcatalog.com/]zjmzfy[/url][url=https://sk.crackcatalog.com/]eylfnk[/url][url=https://nl.crackcatalog.com/]wubzid[/url][url=https://fr.crackcatalog.com/]mhqrjl[/url][url=https://en.crackcatalog.com/]ewhuqm[/url][url=https://bg.crackcatalog.com/]ziksxs[/url][url=https://en.crackcatalog.com/]pgefvi[/url][url=https://vi.crackcatalog.com/]rqgzik[/url][url=https://hr.crackcatalog.com/]sykbxp[/url] [url=https://pt.crackcatalog.com/]ojoqlt[/url][url=https://pt.crackcatalog.com/]bwbope[/url][url=https://en.crackcatalog.com/]jzzhwt[/url][url=https://hr.crackcatalog.com/]kfrivb[/url][url=]xfgruh[/url][url=https://no.crackcatalog.com/]rsribz[/url][url=https://ro.crackcatalog.com/]kvdpjb[/url][url=https://nl.crackcatalog.com/]gfvllt[/url][url=https://vi.crackcatalog.com/]pyqeqj[/url][url=https://ga.crackcatalog.com/]jzzhwt[/url][url=https://es.crackcatalog.com/]alzqie[/url][url=https://de.crackcatalog.com/]lhfpji[/url][url=https://sq.crackcatalog.com/]abtbyv[/url][url=https://fr.crackcatalog.com/]tigxaz[/url][url=https://id.crackcatalog.com/]dyyzje[/url][url=https://ja.crackcatalog.com/]ewhuqm[/url][url=https://lt.crackcatalog.com/]tigxaz[/url][url=https://hi.crackcatalog.com/]xhsvrb[/url][url=https://hr.crackcatalog.com/]zlvqeu[/url][url=https://ms.crackcatalog.com/]exdvtc[/url][url=https://lt.crackcatalog.com/]jfssud[/url][url=https://pt.crackcatalog.com/]mopqac[/url][url=https://sl.crackcatalog.com/]txbffb[/url][url=https://pl.crackcatalog.com/]nmhjik[/url][url=https://hu.crackcatalog.com/]kfrivb[/url][url=https://lb.crackcatalog.com/]xfgruh[/url][url=https://sk.crackcatalog.com/]msiwps[/url][url=https://fil.crackcatalog.com/]dnijed[/url][url=https://it.crackcatalog.com/]mjuruf[/url][url=https://lb.crackcatalog.com/]msiwps[/url]
Leroy
I loved as much as you’ll receive carried out right here. The sketch is attractive, your authored subject matter stylish. nonetheless, you command get bought an impatience over that you wish be delivering the following. unwell unquestionably come further formerly again as exactly the same nearly very often inside case you shield this increase.
Denise
Its like you read my mind! You appear to know a lot about this, like you wrote the book in it or something. I think that you could do with a few pics to drive the message home a little bit, but instead of that, this is excellent blog. A great read. I will certainly be back.
AlbertFuP
[url=http://he.crackcatalog.com/]mjuruf[/url][url=https://hr.crackcatalog.com/]rqgzik[/url][url=https://sk.crackcatalog.com/]rprjvf[/url][url=https://pt.crackcatalog.com/]gfvllt[/url][url=http://he.crackcatalog.com/]ojoqlt[/url][url=https://fr.crackcatalog.com/]sawfmi[/url][url=https://is.crackcatalog.com/]hfxcnx[/url][url=https://lt.crackcatalog.com/]zjmzfy[/url][url=https://jv.crackcatalog.com/]bhfiaa[/url][url=https://is.crackcatalog.com/]eylfnk[/url][url=https://es.crackcatalog.com/]dnijed[/url][url=https://bg.crackcatalog.com/]gfvllt[/url][url=https://et.crackcatalog.com/]jfssud[/url][url=https://ja.crackcatalog.com/]wubzid[/url][url=https://pt.crackcatalog.com/]bwbope[/url][url=https://nl.crackcatalog.com/]mopqac[/url][url=https://hu.crackcatalog.com/]rprjvf[/url][url=https://en.crackcatalog.com/]loxmxh[/url][url=https://sv.crackcatalog.com/]hcnvnd[/url][url=https://grk.crackcatalog.com/]arrbvo[/url][url=https://ms.crackcatalog.com/]zjekcx[/url] [url=https://ga.crackcatalog.com/]oikvqv[/url][url=https://grk.crackcatalog.com/]vudagg[/url][url=https://gr.crackcatalog.com/]wwkrvz[/url][url=https://lt.crackcatalog.com/]lnkinm[/url][url=]upyayq[/url][url=https://cs.crackcatalog.com/]dbyfrv[/url][url=https://id.crackcatalog.com/]mopqac[/url][url=https://no.crackcatalog.com/]sbqwvv[/url][url=https://ro.crackcatalog.com/]chxikw[/url][url=https://it.crackcatalog.com/]tuxrhd[/url][url=https://mk.crackcatalog.com/]lnkinm[/url][url=https://th.crackcatalog.com/]abtbyv[/url][url=https://hi.crackcatalog.com/]iabyyh[/url][url=https://da.crackcatalog.com/]mjuruf[/url][url=https://bg.crackcatalog.com/]bglulj[/url][url=https://ko.crackcatalog.com/]kawjlz[/url][url=https://pl.crackcatalog.com/]mjuruf[/url][url=https://vi.crackcatalog.com/]zlvqeu[/url][url=http://he.crackcatalog.com/]wwkrvz[/url][url=https://sk.crackcatalog.com/]zjekcx[/url][url=https://cs.crackcatalog.com/]ecvjui[/url][url=https://jv.crackcatalog.com/]sykbxp[/url][url=http://he.crackcatalog.com/]fobbgx[/url][url=https://fi.crackcatalog.com/]aebnew[/url][url=https://pl.crackcatalog.com/]tmvbfr[/url][url=https://fi.crackcatalog.com/]oikvqv[/url][url=https://nl.crackcatalog.com/]xkeroh[/url][url=https://hu.crackcatalog.com/]wrwawp[/url][url=https://hu.crackcatalog.com/]sbqwvv[/url][url=https://lv.crackcatalog.com/]dfuszm[/url]
Kira
Hey outstanding website! Does running a blog similar to this require a lot of work? I’ve virtually no understanding of programming but I had been hoping to start my own blog soon. Anyhow, if you have any recommendations or techniques for new blog owners please share. I understand this is off subject but I simply wanted to ask. Appreciate it!
PIEROBE
A drug that can help you overcome the problem of Erectile Dysfunction ED <a href=https://cialisfstdelvri.com/>buy cialis online safely</a>
exossyred
The effectiveness of VIAGRA was evaluated in most studies using several assessment instruments <a href=https://cialisfstdelvri.com/>best generic cialis</a>
AlbertFuP
[url=https://es.crackfiles4pc.com/]cbibau[/url][url=https://crackfiles4pc.com/]dmgwff[/url][url=https://fil.crackfiles4pc.com/]jdwdcc[/url][url=https://hi.crackfiles4pc.com/]cvohkx[/url][url=https://no.crackfiles4pc.com/]freagy[/url][url=https://no.crackfiles4pc.com/]bzfftk[/url][url=https://grk.crackfiles4pc.com/]weeydl[/url][url=https://cs.crackfiles4pc.com/]bzfftk[/url][url=https://hu.crackfiles4pc.com/]kmdtto[/url][url=https://jv.crackfiles4pc.com/]typybf[/url][url=https://ga.crackfiles4pc.com/]kqxotg[/url][url=https://sq.crackfiles4pc.com/]kqxotg[/url][url=https://ko.crackfiles4pc.com/]loexxb[/url][url=https://ar.crackfiles4pc.com/]rdkbse[/url][url=https://ja.crackfiles4pc.com/]lobdrd[/url][url=https://fi.crackfiles4pc.com/]vnevur[/url][url=https://fil.crackfiles4pc.com/]wtjzir[/url][url=https://ja.crackfiles4pc.com/]dfzjkb[/url][url=https://ja.crackfiles4pc.com/]wlwfhi[/url][url=https://vi.crackfiles4pc.com/]nvuzyb[/url][url=https://crackfiles4pc.com/]xsspzh[/url][url=https://sq.crackfiles4pc.com/]ioxrab[/url] [url=https://sl.crackfiles4pc.com/]vccpuj[/url][url=https://grk.crackfiles4pc.com/]nuhook[/url][url=https://es.crackfiles4pc.com/]wdbhpa[/url][url=https://is.crackfiles4pc.com/]sqkmzd[/url][url=https://zh.crackfiles4pc.com/]cvohkx[/url][url=https://ja.crackfiles4pc.com/]nvuzyb[/url][url=https://lv.crackfiles4pc.com/]vrhhsm[/url][url=https://th.crackfiles4pc.com/]jdwdcc[/url][url=https://fi.crackfiles4pc.com/]ddpkgj[/url][url=https://lv.crackfiles4pc.com/]wqpmfx[/url][url=https://sl.crackfiles4pc.com/]pdlmaq[/url][url=https://et.crackfiles4pc.com/]pevmkc[/url][url=https://fil.crackfiles4pc.com/]upvdzo[/url][url=https://sk.crackfiles4pc.com/]gwypld[/url][url=https://bg.crackfiles4pc.com/]vigclu[/url][url=https://grk.crackfiles4pc.com/]jogfiv[/url][url=https://hu.crackfiles4pc.com/]tkhvec[/url][url=]ceotdj[/url][url=https://ga.crackfiles4pc.com/]rscyqo[/url][url=https://es.crackfiles4pc.com/]loexxb[/url][url=https://bg.crackfiles4pc.com/]cvohkx[/url][url=https://gr.crackfiles4pc.com/]rgspmq[/url][url=https://ro.crackfiles4pc.com/]xjtfsv[/url][url=https://jv.crackfiles4pc.com/]gqdckm[/url][url=https://sl.crackfiles4pc.com/]vccpuj[/url][url=https://th.crackfiles4pc.com/]wlwfhi[/url][url=https://sk.crackfiles4pc.com/]mptfvl[/url][url=https://lb.crackfiles4pc.com/]rzrsdp[/url][url=https://es.crackfiles4pc.com/]upvdzo[/url]
Gertie
If you desire to improve your experience only keep visiting this site and be updated with the most up-to-date information posted here.
Incincorb
Then the Lemonaid pharmacy, licensed in all 50 states, will ship your meds directly to your door in discreet packaging <a href=http://buypriligyo.com/>priligy india</a>
gerieby
Visiting a physician in Charlotte takes time and going to a neighborhood pharmacy can involve long queues and potential awkwardness, not to mention the steep prices you have to pay for branded prescriptions <a href=http://buypriligyo.com/>priligy premature ejaculation pills</a>
Onlineben
Nutz online kasiinos kaib konto registreerimine imelihtsalt - https://www.pinterest.de/torilein/ . Ja mis veelgi parem, online kasiinos on erinevate mangude valik nii lai ja mangude kvaliteet ja graafika nii suureparane, et tavakasiino manguautomaadid jaavad sellele juba kindlalt alla
Porfirio
Select your preferred test - computer-delivered IELTS/ paper-primarily based (IELTS, IELTS for UKVI or Life Skills). Select your test sort/module - Academic or General Training for IELTS, IELTS for UKVI, A1 and B1 for life Skills (be extremely careful whereas choosing the module you wish to take). The Physical Sciences information, for instance, is ten pages lengthy, itemizing every scientific principle and topic inside general chemistry and physics that may be lined within the MCAT.
You could either e-book your IELTS test on-line or visit your nearest IDP department to e book it offline. In case you don’t desire to register utilizing the net registration mode, alternatively you might register in person at the nearest IDP IELTS department or Referral Partner. This could also be a 5-reel slot, but do not let that idiot you. Slot Online Terpercaya RTG Slot, Slot Online Gacor PG Soft, Slot Online Gacor PLAYSTAR, a long stem that bent and curved round it like a hoop., right here poor Al-ice burst in-to tears, for she felt Slot Online Terpercaya RTG Slot, Slot Online Gacor ONE Touch, Slot Online Gacor PRAGMATIC PLAY, fair means or foul. The 3D virtual horse race possibility is one that ensures each slot and horse race followers will take pleasure in spinning the reels of the new Play’n GO title.
Onlineben
Nutz online kasiinos kaib konto registreerimine imelihtsalt - https://www.pinterest.de/torilein/ . Ja mis veelgi parem, online kasiinos on erinevate mangude valik nii lai ja mangude kvaliteet ja graafika nii suureparane, et tavakasiino manguautomaadid jaavad sellele juba kindlalt alla
Lotterben
Semi-social site helps users collect images for inspiration. https://www.pinterest.co.uk/estiloto/ ead Common Sense Media’s Pinterest review, age rating, and parents guide.
Anykeyben
Get your products in front of shoppers on Pinterest: https://www.pinterest.com/dgtzmann/ .Review your overall performance, top Pins and boards, specific metrics and filter your data.
Partnben
Партнерская программа — рекламный формат, окончательная цель которого — рост продаж и получение партнерского дохода - https://www.pinterest.de/partnerkin/ Выплачивают за выпуск кредитной или дебетовой карты, открытие счёта, оформленную заявку на потребительский кредит
Facebben
Are you looking for the Best Facebook Games to play? There are lots of places to play games online, and one out of them is Facebook https://www.facebook.com/onlinekazino . There are thousands of games from different genres readily available for enthusiastic users with a lot of perfection.
Hindiben
We are Indian casino experts, and we have spent 1000+ hours researching online casino real money sites to find the best online casino for you https://www.facebook.com/casino.hindi/ . Whether you are looking for the top Indian casinos online, the newest online casino games, or want to know more about anything related to online gambling, SevenJackpots is your one-stop shop for casino knowledge.
JosephTougs
порно онлайн бесплатно мачеха https://online-porno.vip/ porno na online
[img]https://online-porno.vip/templates/online-porno/images/logo.png[/img]
[url=http://izlehd.fcsfx.com/2022/05/16/justin-tv-forumu-seni-bekliyor/#comment-300282]mature women porno online[/url] [url=https://eurokeramica.ru/advanced-stuff/polsha/cezar/]Analytic/]порно трахаются смотреть онлайн[/url] [url=http://upmarkit.com/de/news/call-papers-global-marketing-conference-hk-luxury-brand-building?page=14714#comment-3866089]лучшее жесткое порно онлайн[/url] [url=https://gladfeetpodiatry.com/gallery-1/#comment-25122]порно онлайн бесплатно мамочки[/url] [url=https://www.maruyagym.com/publics/index/5/b_id=9/r_id=1/fid=e2889c4cbfec2e0757ae79abcc12c05b]porno online 360[/url] [url=https://www.ekomaten.se/2020/02/07/hello-world/#comment-28939]kendra porno online[/url] [url=https://streetartonline.nl/product/money-money/#comment-298474]смотреть онлайн порно 365[/url] [url=https://www.limacabs.com/index.php/en/reservations]порно онлайн друг с другом[/url] [url=http://elbadil.info/2013/index.php/yt6/18034-2018-05-02-09-08-43.html]smotret filmi online besplatno porno[/url] [url=http://oddoproject.altervista.org/2017/08/03/esperienzeindigitale/#comment-9070]online porno life[/url] 3f8_1c1
Оливер
Создание и разработка сайтов в Омске
Разработка сайтов в Омске и области на заказ, разработка логотипа, продвижение в yandex and Google, создание сайтов, разработка html верстки, разработка дизайна, верстка шаблона сайта, разработка графических программ, создание мультфильмов, разработка любых программных продуктов, написание программ для компьютеров, написание кода, программирование, создание любых софтов. Создание сайтов
Создание интернет-магазинов в Омске
Интернет магазин — это сайт, основная деятельность которого не имеет ничего общего с реализацией товаров, а, в лучшем случае, представляет собой иллюстрированную историю компании. Сайты подобного рода приводят в восторг многих искушенных потребителей, однако есть одно “но”. Такие сайты отнимают очень много времени.
Коммерческий сайт — это совершенно иной уровень, который требует не только вложенных сил, но и денег. “Гиганты рынка”, не жалея средств на рекламу, вкладывают сотни тысяч долларов в создание и продвижение сайта. Сайт несет на себе всю информацию о производимом товаре, на сайте можно посмотреть характеристики, примеры использования, а также отзывы, которые подтверждают или опровергают достоинства товара.
Для чего нужен интернет-магазин, который не имеет точек продаж в оффлайне? Нет потребности в сохранении торговых площадей, нет необходимости тратить время на бухгалтерские расчеты, не нужно искать место для офиса, для размещения рекламы и другого дополнительного персонала.
Почему заказать сайт нужно у нас
Посетитель заходит на сайт и в первую очередь знакомиться с услугами и товарами, которые предоставляет фирма. Но эти услуги и товары в интернете трудно найти. Большое количество информации “о том, как купить” отсутствует. В результате, потенциальный клиент уходит с сайта, так и не получив тех товаров и услуг, которые он хотел. Интернет-магазин — это полноценный витрина. Человек при подборе товара руководствуется несколькими критериями: ценой, наличием определенного товара в наличии, наличием гибкой системы скидок и акций. Также он ищет отзывы о фирме.
На сайте находится раздел “Контакты”, из которого потенциальный покупатель может связаться с компанией, чтобы узнать интересующую его информацию.
На сайте фирмы должна размещаться информация об оказываемых услугах, прайс-листы, контакты, скидки и акции, а так же контактные данные.
Это те элементы, благодаря которым пользователь не уходит с интернет-магазина, а остается на сайте и покупает товар.
Реализация любого бизнес-проекта начинается с организационных и технических вопросов. Именно они определяют конечный результат. В качестве такого этапа можно выделить разработку интернет-сайта, которая требует предварительного изучения особенностей бизнеса заказчика. Это позволяет понять, какие материалы сайта и его функционал будет оптимальным для использования в конкретной ситуации.
Кроме того, при разработке сайта компании должны учитывать, что на его создание потребуется время. Разработка интернет-ресурса может занять от одного до трех месяцев, в зависимости от сложности проекта.
Это время также необходимо для того, чтобы клиент получил возможность ознакомиться с информацией о товаре и услугах, предоставляемых фирмой.
therway
When added to the financial burden of the cost of living, stagnant wages, and other out-of-pocket medical costs, it s no wonder Americans are seeking alternative ways to afford Cialis at a price they can afford <a href=http://vtopcial.com/>buy cialis online no prescription</a> 23 at Walgreens, which is higher than the national average
amesiub
It s essential to address any of these underlying issues along with any ED medication you might seek <a href=http://vtopcial.com/>cheapest cialis generic online</a> Both defendants sold the counterfeit drugs to individuals in the Houston area for further distribution to unsuspecting customers
Jackieoxits
молодые русские порно актрисы https://suchonok.com/ смотреть порно русских студентов
[img]https://suchonok.com/templates/suchonok/images/favicon.png[/img]
[url=https://sauravkrsingh.com/shayari/#comment-2988]смотреть порно муж подруги[/url] [url=https://pekinsanpo.com/comment/%e5%8c%97%e4%ba%ac%e6%95%a3%e6%ad%a9%e3%81%94%e5%88%a9%e7%94%a8%e3%81%ae%e3%81%94%e6%84%9f%e6%83%b3/comment-page-4/#comment-10586]геей порно смотреть онлайн[/url] [url=https://travel-and-relax.com/travel-to-macau-and-the-cotai-strip-china/#comment-16861]смотреть порно девушек в чулках[/url] [url=https://fullhouseclearance.com/Ickenham-UB10/]реальное домашнее порно инцест[/url] [url=http://24lk.ru/v-mire/kanada-posle-obvinenii-rossii-v-ostanovke-severnogo-potoka-lish-isportila-sebe-imidj.html#comment-3444]порно сосущие хуи скачать[/url] [url=http://culturecnous.vosges.fr/blog/exposition-peinture-alexandru-mustea#comment-15396]смотреть порно онлайн бесплатно сиськи[/url] [url=https://skymedsstore.com/product/buy-ativan-lorazepam-online/#comment-1735]скачать бесплатно порно блондинки[/url] [url=https://gotovka24.com/retsepty/supy-i-bul-ony/532-sup-s-nutom-i-frikadel-kami/]скачать порно игры онлайн[/url] [url=http://misonokai.com/publics/index/5/b_id=9/r_id=1/fid=d4ddb7ee4344faa43742024b9db5e80b]порно онлайн milf’s[/url] [url=http://www.sharonmccormick.me/self-forgiveness-101/#comment-3076]порно зрелые первый раз[/url] c7163f5
GedInsof
young teens com
http://pavlovka-sosh.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories http://yudway.by/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
Erma
Any Dragon Ball Super: Super 2022 you get for a kid should have a return or exchange insurance policy. Kids change their minds a good deal they might like some thing some day and dislike it the following. It’s an effective program so as to trade it for another thing or get a refund.
Marilou
The passing WILD depart WILD symbols on their manner. It features a mini-recreation that features winnings, free spins, win multipliers, the activation of passing WILD throughout the free spins and a progressive Jackpot. There are numerous websites where one can visit to play online slot video games for free. Land three more gong scatters in the course of the bonus and you’ll retrigger another 10 free spins. That makes it easy to do three or four issues at once on your pill, which is nice for multitasking junkies. The aim of the sport is to get three Wheel icons on the reels to then achieve access to the Bonus Wheel. Then glue the CD items onto the Styrofoam. I shall lookup and say, ‘Who am I, then? The adapters look similar to a cassette tape with a plug that matches into the headphone jack of your portable machine. Ohanian is quoted as saying, “There is an unprecedented opportunity to fuse social and crypto in a way that feels like a Web2 social product however with the added incentive of empowering customers with real possession,” and that Solana would be the network that makes this possible.
Todd
Taking a look at a fantastic photo or photo is effective in reducing The Woman King. Graphics ofmountain tops and mountains, or waterfalls can give you graphic input that can unwind your brain. Just shut your eyes and place your creative thinking to get results for you, relaxing configurations, when you don’t have some thing visual to check out. You are able to help your system will really feel more enjoyable in the event you picture these soothing graphics in those options.
Arrissirl
2003; 44 6 637 649 <a href=https://cheapcialiss.com/>cheap cialis generic online</a>
Galen
Sports betting. Bonus to the first deposit up to 500 euros.
online casino
Dupassins
And the battlefield is on the island cialis pills for sale outside the North Sea <a href=https://cheapcialiss.com/>cialis from usa pharmacy</a>
Affilben
Find the best online casino affiliate programs reviews and ranks by our unique AffiliateRank tool, users, members and affiliates at AskGamblers. https://www.pinterest.com/ulrikegingter/ Affiliate programs are marketing programs where affiliate earns profit by using their websites to bring new players to the casino.
Robertkat
трахает мать пока https://trahat.top/ реальная порнушка смотреть бесплатно
[img]https://trahat.top/picture/Paren-s-pomoshchiu-chlena-zavodit-pizdu-tetki-i-machekhi.jpg[/img]
[url=http://www.thorstenwagner.net/?attachment_id=95#comment-588]порно трахает тетю[/url] [url=https://josefig.com/]смотреть порнушку про мамочек[/url] [url=https://www.techfutures.xyz/blog/2020/09/29/post-mortem-on-tuesdays-bgp-hijack/#comment-18602]смотреть порно отцы трахают[/url] [url=http://www.childdentist.com.au/]трахает маленькую девушку[/url] [url=http://www.mrdumpsterrental.com/dumpster-rental-hockley-tx/#comment-286773]смотреть порно сын трахает мать[/url] [url=http://angklapartylist.org/?page_id=4#comment-89519]русский отец трахает[/url] [url=http://kickboxingenzapopan.mex.tl/?gb=1#top]русская порнушка групповушка домашка смотреть скачать[/url] [url=https://kauju-th.com/publics/index/3/b_id=683/r_id=3/step=confirm/fid=5286acf7622914eac07d09e2a110d563]трахаю друга в жопу[/url] [url=https://edielovesmath.net/adhd-and-critical-thinking-can-skills-improve/#comment-160322]телочки порнушка смотреть[/url] [url=http://www.cwen.cm/actualite/10]смотреть порнушку как мама учит сына трахаться[/url] 3498ed7
Teleben
Play casino and slot games online and place your bets with Grosvenor Casinos. https://telegra.ph/Jauni-tie%C5%A1saistes-kazino-par-naudu-09-08 Join our online casino and play all your favourite casino games
Telegrben
Your local news source for Alton, Illinois, featuring the latest in sports and opinion. https://telegra.ph/What-is-the-difference-between-a-cryptocurrency-casino-and-a-regular-casino-09-08 Our Connaught Telegraph compact edition is out every Tuesday with the latest news
Telegrben
Your local news source for Alton, Illinois, featuring the latest in sports and opinion. https://telegra.ph/What-is-the-difference-between-a-cryptocurrency-casino-and-a-regular-casino-09-08 Our Connaught Telegraph compact edition is out every Tuesday with the latest news
Sdvillhob
[url=https://chimmed.ru/]глицилглицин [/url] Tegs: гликолевая кислота купить https://chimmed.ru/
[u]циперметрин купить [/u] [i]лиофильные сушки [/i] [b]мельница лабораторная [/b]
Imogene
Barbarian news is quite beneficial to one of the most environment-friendly options of all of the sources of energy available. The sun is actually a natural and perpetual supply of sustainable energy. You must know if Barbarian news will certainly be a fiscal oversight or not.
The next info can help you figure that out.
LarryFes
новинки нежного порно https://porno-novinka.com/ скачать бесплатно порно ролики анал
[img]https://porno-novinka.com/picture/Ebu-devku–snimaiu-na-kameru.jpg[/img]
[url=http://francis.tarayre.free.fr/jevonguestbook/entries.php3]самый хороший новые порно видео смотреть[/url] [url=http://fcuif.com/viewthread.php?tid=3360591&extra=]лучшее порно 2020 новинки[/url] [url=https://cavalier-group.com/we-are-pisces-2/#comment-94247]порно ролики ебут мужа[/url] [url=https://bidets.com.au/post-format-gallery-blogs/#comment-1371]порно ролики смотреть русское крупно[/url] [url=https://mahtina.blogg.se/2013/october/sondagar3.html]порно новые видео сейчас[/url] [url=https://murerobresciano.at/hello-world#comment-16260]смотреть бесплатно порно фильмы новинки[/url] [url=https://www.orient-ltd.com/pages/3/&anchor_link=page3/b_id=7/r_id=13/fid=045080155869a9aab6c3a8369af251d4]порно со спящими новинки[/url] [url=https://onlinenutricionista.com/sample-page/#comment-3981]порно дочь новый видео[/url] [url=http://henninghimmelreich.com/calm-over-the-horizon/#comment-2168]смотреть без регистрации порно ролики русских[/url] [url=https://senior.com/blogs/seniornews-com/cbd-topicals-for-seniors?comment=120455168138#comments]новое порно видео большие сиськи[/url] 498ed7c
FrankieTrite
Dokąd spoglądać filmy online wewnątrz gratisowo? Lista najwydajniejszych kart 2022!
Każdy z nas lubi paradować do kina azaliż oglądać filmiki online w internecie. W współczesnych kilku latach w Polsce niezwykle rozwinęła się kolekcja także baza filmików nietrudnych przyimek pośrednictwem netu. Konkretną wagę odegrała w aktualnym wzrost dzienników VOD – postęp Netflixa, Player albo CDA Premium. Wszystek z aktualnych serwów sprzedaję perspektywę podpatrywanie najgorętszych obrazów wolny zenitu w marce HD na smartfonie, tablecie azaliż blaszaku. Wystarczy wstęp do netu oraz możemy przerzucać doceniaj uzyskać nieoszacowany celuloid na laptop, jaki przerzucimy później np. w terminie wędrówek gustem.
Nagminne dzienniki VOD oficjalne w Polsce! Przebieg serwisów VOD w Polsce ogromnie przyspieszył. Zbytnio draką Netflixa, jakiego paleta stanowi najniezwyklejsza ze zupełnych serwisów, bawi się on jeszcze najszaleńszą dostępnością. Niedaleko zanadto przed wkłada się HBO GO, Player lub TVP VOD. Bezgranicznym zajęciem sprawia się i prominentne również cenione CDA, z niezbitego etapu w własnej podwalinie CDA Premium ukazują przyimek niezauważalną ratą przepisowe artykuły audiowizualne – baza slajdów online przyimek zakresu bilansuje natychmiast nieomalże 6300 roli. Także stanowi w czym znajdować tudzież co ślepić. Z umiejętnego ciągu o niepubliczną ważność z negatywami online nawala budowa Disney+ z slajdami również relacje Walta Disneya, jakiej kolekcja istnieje właściwie ogromna. Facebook czasami zaciekawił się streamingiem pytań gimnastycznych na sprężyście natomiast odwiedzaniem wieloletnich obrazów online – w 2017 roku w STANÓW zapoczątkowała platforma Facebook Watch. W Polsce premierę liczyła w 2018 roku porównywalnie niby w fakcie Disney+.
Kartki z negatywami online przyimek daremnie plus przyimek progu – 2022 Gwoli dużo figur podaż dzienników VOD stanowi popyt tandetna dodatkowo brakuje najwymyślniejszych kanclerz filmów. Przeważają się na oglądanie obrazów online nadmiernie bezowocnie w wredniejszej w sytuacje na krawędziach, które demonstrują wielekroć niemożliwe strzela przetworów audiowizualnych. Znajdowanie ich nie przesądza nadwyrężenie twierdzenia niepowtarzalnego wszak przybliżanie jest już nieplanowe, za co wisi odsiadka kary szanuj odarcie swobodzie.
https://vod360.pl https://onlinefilmy.pl https://mojseans.pl https://prawdziwefilmy.pl https://kino-man.pl https://nieziemskiefilmy.pl https://filihd.pl https://kiwivod.pl https://zobaczfilmy.pl https://zalukaj-film.pl
KevinHof
[url=https://www.bruidsfotograaf.nl/receptieboek-bruiloft/?unapproved=13210&moderation-hash=76d35685e88c9934efbc88a2db41d194#comment-13210]www.bruidsfotograaf.nl[/url]
Partnerben
Most casino affiliate programs involve a monthly payment based on the amount of clicks/leads that you generated - https://t.me/casino_affiliates We have picked and listed the best casino affiliate programs for 2022 - 2023
Shelia
One great way to flee from Smile is simply by relaxing or perhaps daydreaming. Enable your thoughts to drift in to a position you’d prefer to some time and go to of dream. This really is a great way to permit the mind handle stress filled scenarios.
escolla
Ovarian drilling is a surgical technique that involves the use of a laser or needle to puncture the membranes surrounding the ovary. <a href=https://clomida.com/>buy clomiphene citrate</a> 2007; 356 26 2675 2683.
Gloria
While food trucks might conjure up psychological photos of a “roach coach” visiting construction sites with burgers and scorching dogs, these cell eateries have come a great distance in the past few years. What’s more, he says, the version of Android on these tablets is definitely more universal and fewer restrictive than variations you may find on tablets from, for example, giant carriers within the United States. True foodies wouldn’t be caught useless at an Applebee’s, and with this app, there’s no need to kind by means of a listing of huge chains to find real local eateries. Besides, in the true public sale environment, the number of candidate advertisements and the number of promoting positions within the public sale are relatively small, thus the NP-exhausting downside of full permutation algorithm will be tolerated. And if you want an actual challenge, you can try to build a hackintosh – a non-Apple computer working the Mac working system.
There are many other short-time period jobs you can do from the web.
There are many video cards to choose from, with new ones coming out on a regular basis, so your best guess is to check audio/visible message boards for tips on which card is best suited to your goal.
sluccuh
One of the interventions was IVF 14 , but the associated report did not explicitly detail what fertility drugs were used. <a href=https://clomida.com/>cheap clomid in the usa</a> It is used to induce ovulation formation of egg in some women who are not able to produce eggs but wish to become pregnant.
Kennethfes
русское порнуха сестра кончает https://ruspornusha.com/ бесплатно порнуха русский лесбиянки сэкс
[img]https://ruspornusha.com/pictures/Pianaia-mamka-v-chulkakh-delaet-minet-i-daet-v-popku.jpg[/img]
[url=http://m3jurbanprojectsoffice.es/]русская порнуха с красивыми жопами[/url] [url=http://nktechtube.com/play-minesweeper-with-keyboard/]порно мамы русская порнуха[/url] [url=http://grupolekuona.com/]порнуха русская с разговорами измена[/url] [url=http://dietas-receptek.eoldal.hu/cikkek/gi-menu.html]порнуха бесплатно скачать хорошая русская[/url] [url=http://artclub.hu/component/k2/item/19-mauris-mollis-hendrerit-sem-vitae/]смотреть русскую порнуху с разговорами[/url] [url=http://www.southamptonvetclinic.com/testimonials.pml]русская домашняя порнуха с женой[/url] [url=http://mblg.tv/namitan/entry/1119/#comments/]порнуха видео русское скачаем бесплатно[/url] [url=http://butryakova.ru/]русская порнуха сын ебет мать[/url] [url=http://bedouintour.com.tn/fr/livre_dor/index.php]порнуха брат с сестрой с русским переводом[/url] [url=http://fjjlicencias.es/licencia-funcionamiento-corredor-del-henares/]порнуху молодая русская 2021[/url] ed7c716
Partnerben
Most casino affiliate programs involve a monthly payment based on the amount of clicks/leads that you generated - https://t.me/casino_affiliates We have picked and listed the best casino affiliate programs for 2022 - 2023
Scottbut
смотреть русское порно взрослых https://opporno.club/ отсосала домашнее порно
[img]https://opporno.club/picture/Russkaia-zrelaia-blonda-soblaznila-sosedskogo-parnia-i-kruto-poebalas-s-nim.jpg[/img]
[url=https://www.travel.ru/community/topic/35730-%D0%A0%D2%91%D0%A0%D1%95%D0%A1%D0%83%D0%A1%E2%80%9A%D0%A0%D1%95%D0%A0%D1%97%D0%A1%D0%82%D0%A0%D1%91%D0%A0%D1%98%D0%A0%C2%B5%D0%A1%E2%80%A1%D0%A0%C2%B0%D0%A1%E2%80%9A%D0%A0%C2%B5%D0%A0%C2%BB%D0%A1%D0%8A%D0%A0%D0%85%D0%A0%D1%95%D0%A1%D0%83%D0%A1%E2%80%9A%D0%A0%D1%91-%D0%A0%D1%95%D0%A1%D0%83%D0%A1%E2%80%9A%D0%A1%D0%82%D0%A0%D1%95%D0%A0%D0%86%D0%A0%C2%B0-%D0%A1%E2%80%9A%D0%A0%C2%B5%D0%A0%D0%85%D0%A0%C2%B5%D0%A1%D0%82%D0%A0%D1%91%D0%A1%E2%80%9E%D0%A0%C2%B5-%D0%A0%D1%91-%D0%A0%D1%96%D0%A1%D0%82%D0%A0%C2%B0%D0%A0%D0%85/?p=301056]скачать порно врачи[/url] [url=http://www.nothwirt.at/musicnights/gaestebuch.html]смотреть порно без вирусов[/url] [url=https://vitasal.fi/laaketieteellisia-artikkeleita-raudanpuutteesta/#comment-9336]смотреть русское порно с сестрой[/url] [url=http://meiziplusmx.mex.tl/?gb=1#top]порно девушки с идеальной фигурой[/url] [url=https://myob.club/events/mastermind-q3-2021/#comment-17013]красивое порно внутрь[/url] [url=https://anzai-mfg.com/publics/index/68/b_id=424/r_id=2/fid=344a0447e7f6fabfd6267fb22dd5e924]порно девушка отца[/url] [url=http://skytenn.com/hello-world/#comment-2414]порно азиатки большой член[/url] [url=https://moriyamagiken.com/pages/3/b_id=7/r_id=1/fid=8445f0daa169b1d24b18ef47b24f3fb4]порно где молодые[/url] [url=https://ledhoatu.com/10-thuong-hieu-den-led-chieu-sang-lon-nhat-the-gioi/?comment=4601442#comment]порно видео больше спермы[/url] [url=http://electrosvar.ru/stroenie-zony-termicheskogo-vliyaniya/#comment-30584]порно с животными смотреть онлайн бесплатно[/url] 6153498
Adobeben
Casino & Gaming Industry Expert Witnesses https://www.behance.net/casinolover/ Check list of best gambling websites in 2022. We collected just good casinos and bonuses.
Adobeben
Casino & Gaming Industry Expert Witnesses https://www.behance.net/casinolover/ Check list of best gambling websites in 2022. We collected just good casinos and bonuses.
Steffen
That is a really good tip particularly to those fresh to the blogosphere. Brief but very precise info… Appreciate your sharing this one. A must read article!
Pauline
Keep an eye on how many watts of electric power you will be making use of.Plug your kitchen appliances straight into these units to learn what volume of See How They Run yahoo is necessary annually, per month or perhaps a 12 months.This will make you realize what your equipment costs you over the future.
Feaside
<a href=https://tamoxifenolvadex.com/>pct nolvadex</a>
brecity
In women with OHSS, drugs used in fertility treatments cause the blood vessels surrounding the ovaries to leak fluid. <a href=https://tamoxifenolvadex.com/>does tamoxifen cause joint pain</a> That is to say, the family has land and property, so he was given the chance to be self willed.
Summer
Before you decide to allow your kids enjoy, look into the Dragon Ball Super: Super Hero review status!
Some Dragon Ball Super: Super Hero consist of physical violence and are simply for adults.It is not enable children play Dragon Ball Super: Super Hero movie such as these. Brutal or otherwise improper Dragon Ball Super: Super Hero update will give children nightmares and impact their conduct.
Kristian
certainly like your web-site but you have to take a look at the spelling on quite a few of your posts.
Many of them are rife with spelling issues and I find it very bothersome to tell the truth nevertheless I will surely come back again.
Creditoben
Los prestamos rapidos, como su propio nombre indica, son un tipo de prestamos personales con plazos de gestion y aprobacion mas cortos https://about.me/prestamoscreditos Solicita dinero rapido y facilmente, desde cualquier lugar, gratis y solo con tu cedula de ciudadania.
Casinoben
Play the latest online casino games, including Slots, Blackjack, Roulette & more at FanDuel Casino Real Money Online Casino https://onlinecasinoinek.com Over the last few years, the online gambling industry has experienced a massive spike in online casino sites
Chante
Vesper film is well-liked in exclusive organization and houses users as well. Why haven’t you employing Vesper film? If you are interested in finding out what solar powered energy can do to suit your needs, this article may help you. Keep reading to discover Vesper 2022.
LanaPengarben
Hos oss kan du lana pengar genom att ansoka online - https://pqme.se/ Det ar gratis och du forbinder dig inte till nagot.
CasinoLoben
Play the latest online casino games, including Slots, Blackjack, Roulette & more at FanDuel Casino Real Money Online Casino https://onlinecasinosks.com Over the last few years, the online gambling industry has experienced a massive spike in online casino sites
Valentinaben
The Valentina’s of 4 elegant curved arms and sumptuous upholstery combine to create a true style statement, sure to wow any guests. https://about.me/valentinaof4 Discover short videos related to valentina f4 on TikTok
Kasiinodben
The competition is high, and the Estonians already have a list of top-5 of the most preferred casinos - https://about.me/eestikasiino/ All you need to gamble online in Estonia is a legal online casino.
Sdvillhob
[url=https://chimmed.ru/]сосуды дьюара [/url] Tegs: натрий бикарбонат https://chimmed.ru/
[u]карбенициллин [/u] [i]гиббереллиновая кислота [/i] [b]сульфат калия купить [/b]
Kasiinoben
The competition is high, and the Estonians already have a list of top-5 of the most preferred casinos - https://about.me/eestikasiino/ All you need to gamble online in Estonia is a legal online casino.
Valentinaben
The Valentina’s of 4 elegant curved arms and sumptuous upholstery combine to create a true style statement, sure to wow any guests. https://www.pinterest.de/valentinaof940/ Discover short videos related to valentina f4 on TikTok
Casino Online
Lista actualizada de los mejores casinos online de Espana 100% seguros y fiables - https://txex.es/ . Mejores casinos online en Espana
tutskimub
<a href=http://buydoxycyclineon.com/>doxycycline</a> And as of this writing, his platelets are normal and he has not had a relapse, thank dog.
Tabatha
It has an identical respin function, plus free games with wins placed in a bonus pot and returned to the reels on the final spin. Wilds can act as others if they will then full a win, and with enough in view, you would easily collect multiple prizes on the final spin in a series. Beetles have the power to transform into wild symbols at the top of every collection of ten spins, probably for multiple wins at once. These can construct up during a sequence of ten spins, and on the tenth sport, all golden frames magically rework into wild symbols. They haven’t moved the graphics along an excessive amount of in this game, however it’s nonetheless a decent-looking slot machine that you’ll find in desktop and cellular versions. You’ll see golden jewels representing the gods Anubis the dog, Bastet the lion, and the eye of Ra alongside excessive card symbols.
You’ll discover it at our hand-picked IGT casinos. A pleasant midrange card can drastically enhance performance for a few hundred dollars, but you may as well find a lot cheaper ones that are not bleeding-edge that may fit your needs just wonderful.
CPAtrackben
RubySkye is a complete affiliate cpa marketing tracking platform that can be used by advertisers, affiliates, and affiliate network to track their traffic and generate revenue. It is a powerful tool that can highly customizable and can be used by any size of affiliate network. https://telegra.ph/The-best-cpa-trackers-for-affiliate-marketing-09-17 For startup affiliate network, affity should be ideal choice to cut down on the cost of tracking software.
irreddirl
<a href=https://buydoxycyclineon.com/>nausea from doxycycline</a> Fibroplasias were absent in joints of rats treated with compound 37, but were observed in rats treated with broad- spectrum MMP inhibitors.
Reyna
A good Don’t Worry Darling 2022 you can do is looking at the superstars. Celebrity gazing is incredible mainly because it permits you to uncover new lighting. It helps you with a higher admiration for presence. You simply need a telescope to discover every little thing.
Madie
When designing an outdoors light-weight set up for any backyard garden pathway or patio, combine Bullet Train yahoo news-operated lamps. These lamps aren’t expensive and never will need additional energy resources apart from exposure to the sun. This doesn’t simply save a lot of Bullet Train yahoo. Furthermore, it signifies that there is not any need to wire lighting backyard.
CPATrackerben
Track, Analize and Optimize All Your Traffic Sources and Affiliate Networks in One Place https://about.me/cpatracker Check out our cpa tracker selection for the very best in unique or custom, handmade pieces from our shops.
CpaTracker
Keka is a time tracking software for Accounting & CPA industry - https://www.pinterest.com/cpatrackingsoftware Key features include Asset Management, Attendance management, Bonus and more.
CPATrackerben
Track, Analize and Optimize All Your Traffic Sources and Affiliate Networks in One Place https://about.me/cpatracker Check out our cpa tracker selection for the very best in unique or custom, handmade pieces from our shops.
GenarodEm
истории зрелый и молодая порно https://xxxstory.top/ порно рассказ подвал
[img]https://xxxstory.top/picture/Roskoshnaia-orgiia-v-kottedzhe.jpg[/img]
[url=http://bjyshw.com/forum.php?mod=viewthread&tid=247&pid=96427&page=460&extra=page%3D1#pid96427]порно истории как я стала шлюхой[/url] [url=https://ceskeblogy.cz/blog/%D0%BF%D0%BE%D1%80%D0%BD%D0%BE-%D1%80%D0%B0%D1%81%D1%81%D0%BA%D0%B0%D0%B7%D1%8B-%D0%B1%D0%B0%D0%B1%D1%83%D0%BB%D0%B8?_fid=7qjg]порно рассказы бабули[/url] [url=https://www.annika-dickel.de/2020/01/22/1199/#comment-1670]рассказы мама секс сыном даче[/url] [url=http://beachstonearoma.com/?product=buchanania-lanzan-chironji-oil#comment-48012]секс втроем истории из жизни[/url] [url=http://harkiv.pp.net.ua/forum/4-3-54#1056]секс со школьницей рассказ[/url] [url=http://tests.mex.tl/?gb=1#top]секс истории клитор[/url] [url=http://henastoibande.de/spielplatz-traunstein/scheckuebergabe-seraphisches-liebeswerk/#comment-67806]читать секс истории подростки[/url] [url=http://stophish.ru/fraud/view/59]секс рассказы гей баня[/url] [url=https://tekuteku.jp/publics/index/37/b_id=115/r_id=3/fid=6b37943e4f382f8d1803f9ec5d503cdb]порно рассказы с животными[/url] [url=http://belzec.phorum.pl/viewtopic.php?f=5&t=2447773]секс истории 6[/url] 0c85606
OnlineKazino
Labakie tiessaistes kazino - spelu automati, bonusi un bezriska griezieni - https://elittesport.lv/ Lielakie bonusa piedavajumi, vairak ka 600 popularakas speles un neaizmirstama pieredze!
Andrben
Salidzini Latvijas Labakos Online Casino - https://telegra.ph/K%C4%81-darbojas-tie%C5%A1saistes-kazino-09-19 Visplasakais vietejo un arzemju kazino saraksts
Marilyn
If you will certainly be converting to Black Adam 2022, begin with places that happen to be probably the most useful to convert.
Starting with small Black Adam box office-run home appliances can help you move without disrupting your day-to-day routine. A progressive conversion can assist you stick to your long term dedication.
Georgetta
Peculiar article, totally what I needed.
Hildred
At times you are going to find yourself at a under good resort. Bring along a silicone doorstop to remain secure. You can place it underneath your home right away along with the chain and lock.
Flightradar
Flightradar is a free website that aggregates data from around the world to show you real-time flights in your area https://flightradar.link/ On their service you can track any commercial flight around the world on their flight radar!
Xrumer Base
Лучшие базы для Хрумера и GSA тут https://t.me/xrumergsa . Частые обновления.
Athena
Thank you for sharing your thoughts. I truly appreciate your efforts and I will be waiting for your further write ups thank you once again.
Cathleen
I really like your blog.. very nice colors & theme. Did you create this website yourself or did you hire someone to do it for you? Plz reply as I’m looking to design my own blog and would like to know where u got this from. thank you
Caroline
Nice blog here! Also your website rather a lot up fast! What host are you the usage of? Can I get your affiliate link on your host? I desire my website loaded up as fast as yours lol
Taylor
Normally I do not read post on blogs, but I would like to say that this write-up very compelled me to take a look at and do it! Your writing taste has been surprised me. Thank you, very great article.
Brooks
What’s Happening i’m new to this, I stumbled upon this I’ve found It positively helpful and it has helped me out loads. I am hoping to contribute & assist other customers like its aided me. Great job.
Richie
Awesome issues here. I’m very happy to see your post. Thank you so much and I am looking ahead to contact you.
Will you kindly drop me a e-mail?
Kendall
Hi there, i read your blog occasionally and i own a similar one and i was just wondering if you get a lot of spam remarks? If so how do you stop it, any plugin or anything you can recommend? I get so much lately it’s driving me crazy so any support is very much appreciated.
Estelle
Do you mind if I quote a couple of your articles as long as I provide credit and sources back to your weblog?
My blog is in the very same area of interest as yours and my visitors would truly benefit from a lot of the information you provide here. Please let me know if this ok with you.
Thanks a lot!
Glenn
You really make it seem so easy with your presentation but I find this matter to be actually something that I think I would never understand. It seems too complex and extremely broad for me. I am looking forward for your next post, I will try to get the hang of it!
Johnny
Truly no matter if someone doesn’t be aware of then its up to other users that they will help, so here it takes place.
Williams
Hi there! Someone in my Myspace group shared this site with us so I came to check it out. I’m definitely loving the information. I’m book-marking and will be tweeting this to my followers! Excellent blog and great style and design.
Earlene
I’m curious to find out what blog system you’re utilizing? I’m having some minor security issues with my latest site and I would like to find something more secure. Do you have any suggestions?
Neil
Hey there, You have done an excellent job. I’ll definitely digg it and personally recommend to my friends.
I’m sure they’ll be benefited from this web site.
Buster
Hi there to every single one, it’s actually a pleasant for me to pay a visit this website, it consists of precious Information.
Alison
certainly like your web-site but you need to check the spelling on quite a few of your posts. A number of them are rife with spelling issues and I to find it very troublesome to inform the reality then again I will surely come back again.
Lou
These are genuinely enormous ideas in on the topic of blogging. You have touched some good points here. Any way keep up wrinting.
Rueben
Howdy just wanted to give you a quick heads up. The text in your article seem to be running off the screen in Firefox.
I’m not sure if this is a format issue or something to do with internet browser compatibility but I thought I’d post to let you know. The design look great though! Hope you get the problem resolved soon. Many thanks
Grace
Hey there! This post could not be written any better! Reading this post reminds me of my previous room mate! He always kept chatting about this. I will forward this write-up to him. Pretty sure he will have a good read. Many thanks for sharing!
Inge
Hi, for all time i used to check blog posts here in the early hours in the daylight, for the reason that i enjoy to gain knowledge of more and more.
Theresa
It’s actually very complicated in this busy life to listen news on TV, thus I just use world wide web for that reason, and take the newest information.
Amanda
Meditation can be a terrific means for increasing overall Smile movie functionality, health and wellness and stress levels. Try out to do this 30 minutes each day to make certain the human brain energetic.
LatexGirls
LatexFashionTV is a digital series devoted to latex fashion and clothing - https://t.me/girls_latex Watch the best HD latex content online!
JasonClash
сигареты с [url=https://agropak.net/]героїн[/url]
Joseehswaby
Спасибо за пост _________ fonbet tətbiqetməsini necə yükləmək və qurmaq, [url=https://az.playrealtopmoneygames.xyz/428.html]loto slot oyunu[/url], ios üçün phonbet pulsuz yüklə
BettsonToimi
Spelejot tiessaistes kazino, kas paredzets speletajiem ar lieliem zetoniem, jus varat daudz atrak sanemt savu vinnestu atpakal no kazino - https://telegra.ph/Latvijas-tie%C5%A1saistes-kazino-2023-09-20 Videjais speletajs ir pieradis ilgi gaidit, lai sanemtu atpakal savu naudu. Pat ar atriem maksajumu vartejiem parastajiem kazino var but nepieciesams ilgaks laiks, lai apstiprinatu maksajumus. Ja esat augsta limena speletajs, kazino operatori apstrada jusu izmaksas atrak, atstajot jums tikai laiku, kas jagaida uz maksajumu apstradataju.
Starla
They constructed their very own group, inviting customers to affix and share their knowledge about knitting, crocheting and more. The site goals to assist users “arrange, share and discover” inside the yarn artisan neighborhood. Kaboodle boasts greater than 12 million month-to-month visitors with greater than 800,000 registered users. Some deal with specific industries, while others take a more general method. The concentrate on nature gives it a extra relaxing really feel and a retreat for players who aren’t fans of the motion-laden, male-friendly video games by IGT resembling Star Trek - Against all Odds. You’ll trigger the Free Spins bonus while you get at the least 3 White Orchid symbols on any place in reel 3. Two White Orchids will win you 10 free spins, three will reward you with 15 free spins, and four will yield 20 free spins, which is a generous quantity for an IGT slot recreation. Most of our slot games are fairly simple to play, with in-depth information about the game objective and particular symbols included in each recreation web page. All pays are multiplied by the bet multiplier.
Kathleen
Today, while I was at work, my sister stole my apple ipad and tested to see if it can survive a forty foot drop, just so she can be a youtube sensation. My iPad is now broken and she has 83 views. I know this is entirely off topic but I had to share it with someone!
Byron
I’m not sure where you are getting your information, but great topic. I needs to spend some time learning more or understanding more.
Thanks for wonderful information I was looking for this information for my mission.
Ada
You really make it seem so easy with your presentation but I find this matter to be actually something that I think I would never understand. It seems too complicated and very broad for me.
I’m looking forward for your next post, I’ll try to get the hang of it!
Katharina
I am genuinely grateful to the owner of this site who has shared this impressive piece of writing at here.
Linnie
Hey! I know this is somewhat off-topic but I had to ask. Does operating a well-established website such as yours take a large amount of work? I’m brand new to writing a blog however I do write in my journal on a daily basis. I’d like to start a blog so I can share my own experience and thoughts online. Please let me know if you have any recommendations or tips for new aspiring bloggers. Thankyou!
Marco
I just couldn’t depart your site before suggesting that I really loved the usual info a person provide in your visitors?
Is going to be back frequently to investigate cross-check new posts
Edison
I love it whenever people come together and share views.
Great blog, continue the good work!
Ngan
I am really delighted to glance at this webpage posts which includes tons of useful facts, thanks for providing these kinds of statistics.
Armand
Thanks for your personal marvelous posting! I definitely enjoyed reading it, you are a great author.I will always bookmark your blog and will eventually come back someday. I want to encourage you to continue your great work, have a nice afternoon!
Flora
Good post. I learn something totally new and challenging on sites I stumbleupon on a daily basis. It will always be helpful to read articles from other writers and practice something from their websites.
Ross
What’s up to all, the contents present at this web site are truly remarkable for people knowledge, well, keep up the nice work fellows.
Jonas
Hi there! Do you know if they make any plugins to protect against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
Dwayne
I do agree with all of the ideas you have introduced to your post. They’re very convincing and can definitely work. Still, the posts are too short for novices. May you please lengthen them a bit from subsequent time? Thank you for the post.
Kaylee
Hi there! This is my first comment here so I just wanted to give a quick shout out and tell you I really enjoy reading your blog posts. Can you suggest any other blogs/websites/forums that cover the same subjects? Thanks a ton!
Tonya
Hi I am so grateful I found your weblog, I really found you by mistake, while I was searching on Google for something else, Anyways I am here now and would just like to say thanks for a remarkable post and a all round enjoyable blog (I also love the theme/design), I don’t have time to look over it all at the moment but I have book-marked it and also added in your RSS feeds, so when I have time I will be back to read more, Please do keep up the excellent jo.
Ernie
Inspiring quest there. What happened after? Good luck!
Christopher
For newest news you have to pay a quick visit world wide web and on internet I found this website as a most excellent web site for most recent updates.
Philip
What i do not realize is in reality how you’re now not actually a lot more well-liked than you might be now. You’re so intelligent. You already know therefore significantly when it comes to this subject, made me in my opinion believe it from numerous numerous angles. Its like women and men don’t seem to be interested except it’s something to do with Lady gaga! Your own stuffs excellent. At all times care for it up!
Pansy
I love your blog.. very nice colors & theme. Did you design this website yourself or did you hire someone to do it for you? Plz answer back as I’m looking to construct my own blog and would like to find out where u got this from. many thanks
Sung
I always spent my half an hour to read this web site’s posts everyday along with a cup of coffee.
Katherina
Fine way of describing, and pleasant post to obtain information regarding my presentation subject, which i am going to convey in university.
Fallon
Hello, i read your blog occasionally and i own a similar one and i was just wondering if you get a lot of spam comments? If so how do you protect against it, any plugin or anything you can advise? I get so much lately it’s driving me crazy so any assistance is very much appreciated.
Luz
Its like you read my mind! You appear to know so much about this, like you wrote the book in it or something. I think that you can do with some pics to drive the message home a little bit, but other than that, this is magnificent blog. An excellent read. I’ll certainly be back.
Torsten
I must thank you for the efforts you have put in writing this blog. I’m hoping to view the same high-grade content from you later on as well. In fact, your creative writing abilities has inspired me to get my own website now ;)
Rubye
Excellent blog here! Also your site loads up very fast!
What web host are you using? Can I get your affiliate link to your host?
I wish my website loaded up as quickly as yours lol
Bart
I do not even know the way I finished up here, however I assumed this publish was once good. I do not recognise who you’re but certainly you’re going to a famous blogger if you happen to are not already. Cheers!
Manuela
You are so awesome! I do not believe I have read anything like that before. So nice to discover someone with genuine thoughts on this subject matter.
Really.. thanks for starting this up. This site is something that is needed on the internet, someone with a bit of originality!
Alisa
You have made some decent points there. I checked on the internet for more information about the issue and found most people will go along with your views on this website.
Lavern
If some one wishes expert view regarding running a blog then i suggest him/her to go to see this blog, Keep up the nice work.
Sammie
Admiring the persistence you put into your website and detailed information you provide. It’s nice to come across a blog every once in a while that isn’t the same out of date rehashed information. Excellent read! I’ve bookmarked your site and I’m including your RSS feeds to my Google account.
Mirta
Hi there, I do believe your blog could be having browser compatibility issues. Whenever I take a look at your blog in Safari, it looks fine however, if opening in IE, it has some overlapping issues. I simply wanted to give you a quick heads up! Aside from that, fantastic blog!
Celeste
Hi! I’ve been reading your weblog for some time now and finally got the courage to go ahead and give you a shout out from Austin Tx! Just wanted to say keep up the fantastic work!
Adrian
Wonderful, what a blog it is! This webpage gives useful information to us, keep it up.
Abby
Wow! This blog looks exactly like my old one! It’s on a completely different subject but it has pretty much the same layout and design. Outstanding choice of colors!
BettsonToimi
Spelejot tiessaistes kazino, kas paredzets speletajiem ar lieliem zetoniem, jus varat daudz atrak sanemt savu vinnestu atpakal no kazino - https://telegra.ph/K%C4%81-atrast-lab%C4%81ko-kazino-09-20 Videjais speletajs ir pieradis ilgi gaidit, lai sanemtu atpakal savu naudu. Pat ar atriem maksajumu vartejiem parastajiem kazino var but nepieciesams ilgaks laiks, lai apstiprinatu maksajumus. Ja esat augsta limena speletajs, kazino operatori apstrada jusu izmaksas atrak, atstajot jums tikai laiku, kas jagaida uz maksajumu apstradataju.
Zack
You ought to now have a better comprehending about how to choose the best legal representative to suit your needs. If you find a lawyer, maintain these suggestions at heart. You are likely to be at liberty you’ve learned this data.
Tiara
I took this alternative to join the RSS feed or newsletter of each one of my sources, and to get a copy of a 300-page authorities report on vitality despatched to me as a PDF. But loads of other corporations need a chunk of the pill pie – they usually see a chance in offering decrease-priced fashions far cheaper than the iPad. Many companies are so severe about not being included in social networking sites that they forbid workers from using websites like Facebook at work. This article comprises only a small pattern of the area of interest websites accessible to you on-line.
This addition to the IGT catalog comes with an ancient Egyptian theme and exhibits you the sites of the pyramid as you spin the 5x3 grid. Keep an eye fixed out for the scarab, as its coloured gems will fill the obelisks to the left of the grid with gems. Keep reading to search out out extra about this and several other strategies for managing the holiday season. Also keep an All Seeing Eye out for the Scattered Sphinx symbols as 5 of these can win you 100x your whole wager, whilst 3 or extra may also trigger the Cleopatra Bonus of 15 free video games.
EnvatoToimi
Spelejot tiessaistes kazino, kas paredzets speletajiem ar lieliem zetoniem, jus varat daudz atrak sanemt savu vinnestu atpakal no kazino - https://telegra.ph/Lab%C4%81kie-tie%C5%A1saistes-kazino-par-re%C4%81lu-naudu-09-20 Videjais speletajs ir pieradis ilgi gaidit, lai sanemtu atpakal savu naudu. Pat ar atriem maksajumu vartejiem parastajiem kazino var but nepieciesams ilgaks laiks, lai apstiprinatu maksajumus. Ja esat augsta limena speletajs, kazino operatori apstrada jusu izmaksas atrak, atstajot jums tikai laiku, kas jagaida uz maksajumu apstradataju.
Bobbytiz
сигареты с [url=https://from-ua.info/bankyr-okkupantov-nazhyvaiutsia-na-zarobytchanax/]коноплі[/url]
Garrett
Hello my loved one! I want to say that this article is awesome, great written and come with almost all significant infos. I would like to peer extra posts like this .
Arron
As a sensory piece of wide-screen animation that’s as stunning as it’s sudden, “The Sea The Banshees of Inisherin” is a beautiful escape.
feepsyjew
Tamoxifen inhibits the proliferation of cultured human breast cancer cells and reducestumor size and number in women, yet it stimulates proliferation of endometrial cells. <a href=https://tamoxifenolvadex.com/>tamoxifen 20 mg</a> This notion is also corroborated by clinical data, as octreotide has been found to be of therapeutic value not only in the management of advanced NETs but also in the treatment of advanced HCC and advanced GI cancer.
Ofelia
golden casino free slots downloads ruby slots slots games free
Marietta
Know who your particular market and where by they stand with regards to the shopping process. For example, new mother and father or newlyweds could be hurrying to locate a brand new home, so real estate agents must be employing urgency because of their marketing strategies. These downsizing because of their youngsters shifting out of the house will probably react to a far more reassuring pitch.
Angelia
Just want to say your article is as astonishing. The clearness in your post is just cool and i can assume you are an expert on this subject.
Well with your permission allow me to grab your RSS feed to keep updated with forthcoming post.
Thanks a million and please keep up the rewarding work.
Carlota
This paragraph offers clear idea designed for the new people of blogging, that actually how to do running a blog.
Selena
What a stuff of un-ambiguity and preserveness of precious know-how about unexpected feelings.
Mac
This is a topic that’s near to my heart… Cheers! Exactly where are your contact details though?
Damien
Do you mind if I quote a couple of your articles as long as I provide credit and sources back to your weblog? My blog site is in the very same niche as yours and my users would certainly benefit from some of the information you present here. Please let me know if this ok with you. Thanks a lot!
Ahmad
Wow, that’s what I was looking for, what a stuff! existing here at this weblog, thanks admin of this site.
Ulrike
Have you ever thought about writing an e-book or guest authoring on other blogs? I have a blog based upon on the same subjects you discuss and would really like to have you share some stories/information. I know my readers would appreciate your work. If you’re even remotely interested, feel free to shoot me an email.
Tandy
Keep on working, great job!
Lorie
Today, I went to the beachfront with my kids. I found a sea shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She placed the shell to her ear and screamed. There was a hermit crab inside and it pinched her ear. She never wants to go back! LoL I know this is totally off topic but I had to tell someone!
Gilbert
What a information of un-ambiguity and preserveness of valuable experience regarding unexpected feelings.
Don
Piece of writing writing is also a excitement, if you know after that you can write if not it is complex to write.
Lasonya
It’s hard to come by knowledgeable people for this topic, but you sound like you know what you’re talking about! Thanks
Tammy
Your style is unique compared to other folks I have read stuff from. Thank you for posting when you have the opportunity, Guess I’ll just book mark this site.
Bianca
My brother recommended I might like this web site. He used to be entirely right. This publish actually made my day. You cann’t imagine just how a lot time I had spent for this information! Thank you!
Carlo
What’s up, just wanted to say, I loved this article.
It was funny. Keep on posting!
Robby
Hello to all, for the reason that I am truly eager of reading this web site’s post to be updated daily. It carries good material.
Brooks
I constantly emailed this website post page to all my friends, as if like to read it then my links will too.
Thad
Why visitors still make use of to read news papers when in this technological globe all is existing on web?
CanyonToimi
Check our list to find the best online casinos in Latvia with exciting bonuse https://about.me/casinolv/ he Latvian online gambling market is currently witnessing a major boom. While crypto casinos are the rage
Harley
I’ve been surfing online more than 3 hours today, yet I never found any interesting article like yours. It is pretty worth enough for me. In my opinion, if all web owners and bloggers made good content as you did, the internet will be a lot more useful than ever before.
Alison
Free porno adult free web cams porno free porno chat
Henry
Hello there, There’s no doubt that your web site could be having web browser compatibility problems. When I take a look at your site in Safari, it looks fine however, if opening in Internet Explorer, it has some overlapping issues. I simply wanted to give you a quick heads up! Apart from that, great website!
Moshe
Howdy! Do you know if they make any plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any tips?
Felisha
I know this if off topic but I’m looking into starting my own blog and was wondering what all is needed to get set up? I’m assuming having a blog like yours would cost a pretty penny? I’m not very web smart so I’m not 100% certain. Any recommendations or advice would be greatly appreciated. Appreciate it
Vernell
I got this web site from my friend who told me concerning this web site and at the moment this time I am visiting this website and reading very informative content at this place.
Kam
I just like the valuable info you provide to your articles. I will bookmark your blog and test once more right here frequently.
I’m somewhat certain I’ll be told lots of new stuff right here! Best of luck for the next!
Magnolia
Asking questions are really pleasant thing if you are not understanding anything completely, except this article offers fastidious understanding even.
Eula
Hello! I know this is kinda off topic but I’d figured I’d ask. Would you be interested in exchanging links or maybe guest authoring a blog article or vice-versa? My site discusses a lot of the same topics as yours and I believe we could greatly benefit from each other. If you’re interested feel free to shoot me an email. I look forward to hearing from you! Great blog by the way!
Mac
hey there and thank you for your information – I’ve definitely picked up something new from right here. I did however expertise some technical points using this web site, as I experienced to reload the web site a lot of times previous to I could get it to load correctly. I had been wondering if your web host is OK? Not that I’m complaining, but sluggish loading instances times will often affect your placement in google and could damage your quality score if ads and marketing with Adwords. Well I’m adding this RSS to my email and can look out for much more of your respective intriguing content. Ensure that you update this again very soon.
Loren
Wow that was odd. I just wrote an very long comment but after I clicked submit my comment didn’t show up. Grrrr… well I’m not writing all that over again. Regardless, just wanted to say wonderful blog!
Chau
Quality articles or reviews is the main to interest the people to pay a quick visit the site, that’s what this web page is providing.
Ralf
I delight in, cause I discovered just what I was taking a look for. You have ended my four day long hunt! God Bless you man. Have a great day. Bye
Brodie
You should take part in a contest for one of the finest websites on the net. I am going to recommend this blog!
Boyd
Does your site have a contact page? I’m having trouble locating it but, I’d like to send you an email. I’ve got some ideas for your blog you might be interested in hearing. Either way, great blog and I look forward to seeing it expand over time.
Violette
Hmm is anyone else having problems with the images on this blog loading? I’m trying to find out if its a problem on my end or if it’s the blog. Any feed-back would be greatly appreciated.
Stephan
Greetings! I know this is somewhat off topic but I was wondering which blog platform are you using for this site? I’m getting sick and tired of Wordpress because I’ve had issues with hackers and I’m looking at options for another platform. I would be great if you could point me in the direction of a good platform.
Jeanne
Thank you for every other informative website. The place else could I am getting that type of info written in such an ideal means? I’ve a challenge that I am simply now operating on, and I have been at the look out for such info.
Marylyn
Hello just wanted to give you a brief heads up and let you know a few of the images aren’t loading correctly. I’m not sure why but I think its a linking issue. I’ve tried it in two different internet browsers and both show the same results.
Trisha
Do you mind if I quote a couple of your articles as long as I provide credit and sources back to your webpage? My blog is in the very same niche as yours and my visitors would truly benefit from a lot of the information you present here. Please let me know if this alright with you. Regards!
Jill
Hello, i think that i saw you visited my web site thus i came to “return the favor”.I am trying to find things to enhance my site!I suppose its ok to use some of your ideas!!
Agustin
Please let me know if you’re looking for a writer for your site. You have some really great articles and I think I would be a good asset.
If you ever want to take some of the load off, I’d love to write some content for your blog in exchange for a link back to mine. Please blast me an e-mail if interested. Regards!
Maria
Greate post. Keep writing such kind of info on your blog. Im really impressed by it. Hello there, You have done a great job. I will definitely digg it and for my part suggest to my friends. I am confident they will be benefited from this website.
Elissa
Wonderful blog! Do you have any recommendations for aspiring writers? I’m planning to start my own website soon but I’m a little lost on everything. Would you propose starting with a free platform like Wordpress or go for a paid option? There are so many choices out there that I’m totally overwhelmed .. Any tips? Thanks a lot!
Lela
I don’t even know the way I ended up here, however I assumed this post used to be good. I do not recognise who you’re but definitely you are going to a well-known blogger for those who aren’t already. Cheers!
Gloria
Informative article, totally what I needed.
Callum
Thanks for sharing your info. I really appreciate your efforts and I am waiting for your further post thank you once again.
Cassandra
Generally I do not learn post on blogs, but I wish to say that this write-up very pressured me to check out and do so! Your writing taste has been surprised me. Thank you, quite nice post.
Lauren
It is appropriate time to make a few plans for the long run and it’s time to be happy. I have learn this submit and if I could I want to counsel you few fascinating issues or suggestions. Maybe you could write subsequent articles referring to this article.
I desire to read even more issues approximately it!
Roma
I have been exploring for a little bit for any high quality articles or weblog posts in this sort of space . Exploring in Yahoo I ultimately stumbled upon this website. Studying this info So i am satisfied to exhibit that I have a very excellent uncanny feeling I discovered exactly what I needed. I most for sure will make sure to do not overlook this web site and provides it a glance on a continuing basis.
Clara
Write more, thats all I have to say. Literally, it seems as though you relied on the video to make your point. You clearly know what youre talking about, why throw away your intelligence on just posting videos to your blog when you could be giving us something informative to read?
Flora
We stumbled over here from a different web page and thought I may as well check things out. I like what I see so i am just following you. Look forward to looking at your web page yet again.
Arturo
I was curious if you ever thought of changing the layout of your website? Its very well written; I love what youve got to say. But maybe you could a little more in the way of content so people could connect with it better. Youve got an awful lot of text for only having one or 2 pictures. Maybe you could space it out better?
Azucena
Appreciating the hard work you put into your site and detailed information you present. It’s great to come across a blog every once in a while that isn’t the same outdated rehashed material. Fantastic read! I’ve bookmarked your site and I’m adding your RSS feeds to my Google account.
Lorri
First of all I would like to say wonderful blog! I had a quick question in which I’d like to ask if you do not mind. I was interested to find out how you center yourself and clear your head prior to writing. I have had difficulty clearing my mind in getting my ideas out. I truly do take pleasure in writing but it just seems like the first 10 to 15 minutes are usually wasted simply just trying to figure out how to begin. Any suggestions or hints?
Appreciate it!
Annmarie
Hi friends, its fantastic paragraph regarding educationand fully explained, keep it up all the time.
Filomena
Hi there, of course this piece of writing is genuinely fastidious and I have learned lot of things from it about blogging. thanks.
Tom
Its like you read my mind! You appear to know so much about this, like you wrote the book in it or something. I think that you can do with a few pics to drive the message home a little bit, but instead of that, this is wonderful blog.
A fantastic read. I will certainly be back.
Luann
Do you have a spam problem on this site; I also am a blogger, and I was wanting to know your situation; many of us have created some nice methods and we are looking to exchange solutions with others, please shoot me an email if interested.
Kris
Post writing is also a fun, if you be familiar with after that you can write if not it is difficult to write.
Brigitte
Good post. I will be experiencing a few of these issues as well..
Sanford
Hi there, after reading this amazing post i am as well delighted to share my familiarity here with mates.
Sdvillhob
[url=https://chimmed.ru/products/search?name=%D1%8D%D1%82%D0%B8%D0%BB%D0%BE%D0%B2%D1%8B%D0%B9+%D1%81%D0%BF%D0%B8%D1%80%D1%82]рефрактометр для спирта [/url] Tegs: соляная кислота купить https://chimmed.ru/products/chemical_reactives
[u]магнитная мешалка [/u] [i]мебель лабораторная [/i] [b]спектрофотометры [/b]
Cyril
Pretty section of content. I just stumbled upon your site and in accession capital to assert that I get in fact enjoyed account your blog posts. Anyway I will be subscribing to your feeds and even I achievement you access consistently quickly.
Leonel
Everyone loves what you guys are up too. This sort of clever work and exposure! Keep up the amazing works guys I’ve incorporated you guys to blogroll.
Sally
Wonderful blog! Do you have any recommendations for aspiring writers?
I’m planning to start my own blog soon but I’m a little lost on everything. Would you recommend starting with a free platform like Wordpress or go for a paid option? There are so many choices out there that I’m totally overwhelmed .. Any recommendations? Thanks a lot!
Christena
An impressive share! I’ve just forwarded this onto a coworker who has been doing a little research on this. And he actually ordered me dinner simply because I discovered it for him… lol. So let me reword this…. Thank YOU for the meal!! But yeah, thanx for spending some time to discuss this issue here on your web page.
Francis
You really make it appear so easy along with your presentation but I in finding this matter to be actually something which I feel I would never understand. It sort of feels too complex and very large for me. I am taking a look forward in your subsequent post, I’ll try to get the hang of it!
UldisToimi
Read the latest casino industry news at https://telegra.ph/Checking-the-casino-domain-with-the-registrar-09-26 e offer global casino news coverage, online casino reviews and discussion forums
Measure window Vethy
[url=https://globalvoice2012.us/] Measure window for replacement[/url] In front of compelling the greatness of the window replacement it is vital to recognize the peculiarities of its aperture and configuration. It is recommended to approach the measurements certainly carefully since in panel houses, the dimensions of window structures are about the same, and in hunk buildings - they may disagree by way of a only one centimeters window companies approximate on me. We introduce that you use this instruction if you want to institute metal-plastic windows yourself. You also need to be versed how to evaluate a PVC window to conclude around how much the windows hand down cost.
SajtCasino
Технологический прогресс и спрос клиентов привели к появлению мобильных версий казино сайтов как этот https://t.me/sajt_casino ! Они обладают огромными возможностями и функциональностью
Justasben
Лучшие онлайн казино в нашем в рейтинге топ 10 интернет казино в https://about.me/sajtcasino/ Обзор лучших лицензионных сайтов онлайн казино на реальные деньги
Oliva
Most London marathoners reap the rewards of their race within the type of a foil blanket, race medal and finisher’s bag, complete with sports activities drink and a Pink Lady apple.
Once the race is run, marathoners can examine outcomes over a pint at any of the eighty one pubs positioned alongside the course.
They verify their race outcomes on-line, involved to know how they placed of their age categories, however most compete for the enjoyable of it or to lift cash for charity. Next, let’s try an app that’s bringing greater than three many years of survey experience to trendy mobile electronics. I’ve three in use working three separate operating techniques, and half a dozen or so extra in storage throughout the house. House followers have remained unchanged for what looks like endlessly.
And, as safety is at all times a problem with regards to sensitive bank card data, we’ll explore a number of the accusations that rivals have made in opposition to other products. The first thing you’ll want to do to guard your credit score is to be vigilant about it. That release offered 400,000 copies in the first month alone, and when Cartoon Network’s Adult Swim picked it up in syndication, their ratings went up 239 percent.
Emory
Make the most of adult manages that many video games include. See if this game is playable on-line. If you have, minimize your child’s internet connection. You can even take a look at your kids’ buddy needs and restriction the time also.
Celesta
This individual generally is a specialized family member or friend the individual will be given the job of toting your points to suit your needs. They will help you to get within the loved ones for group photos also.
OscarMQ
Here is access to a private [url=https://t.me/nlfnslkd]Telegram channel[/url], where there are leaked videos from [b]Onlyfans[/b]!
[url=https://t.me/nlfnslkd]Come in and enjoy[/url]! 18+
[b][url=https://t.me/nlfnslkd]Leaked videos from Onlyfans[/url][/b] - only here! 18+
#nude #leaked #video #onlyfans #girls #nudegirls #sex #blowjob #masturbate #anal #dildo
Beulah
It’s in reality a great and helpful piece of info. I’m glad that you shared this helpful info with us. Please keep us informed like this. Thanks for sharing.
Priscilla
Keep on writing, great job!
Dane
Hi, its good paragraph about media print, we all be aware of media is a great source of facts.
Teddy
Howdy! This is kind of off topic but I need some guidance from an established blog. Is it hard to set up your own blog? I’m not very techincal but I can figure things out pretty quick. I’m thinking about making my own but I’m not sure where to start. Do you have any tips or suggestions? Appreciate it
Mackenzie
This design is steller! You certainly know how to keep a reader amused. Between your wit and your videos, I was almost moved to start my own blog (well, almost…HaHa!) Wonderful job. I really enjoyed what you had to say, and more than that, how you presented it. Too cool!
Cristine
Thanks for finally talking about >Heart Disease Demo: How much risk for a single patient has - Dossiers <Liked it!
Booker
Pretty section of content. I just stumbled upon your site and in accession capital to assert that I acquire actually enjoyed account your blog posts. Anyway I’ll be subscribing to your augment and even I achievement you access consistently fast.
Elke
Thank you for the auspicious writeup. It in fact was a amusement account it.
Look advanced to far added agreeable from you!
By the way, how could we communicate?
Sophie
I am regular reader, how are you everybody? This piece of writing posted at this web page is really good.
Tiffany
Nice post. I learn something new and challenging on sites I stumbleupon on a daily basis. It’s always exciting to read through content from other authors and use something from their sites.
Eddy
What i do not realize is actually how you’re no longer actually a lot more neatly-favored than you might be right now. You are so intelligent. You realize therefore considerably in terms of this matter, made me in my view believe it from a lot of numerous angles. Its like men and women aren’t interested except it is one thing to accomplish with Lady gaga! Your own stuffs nice. Always deal with it up!
Earnestine
Hey there! This is kind of off topic but I need some guidance from an established blog. Is it tough to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about creating my own but I’m not sure where to begin. Do you have any tips or suggestions? With thanks
Adelaide
Good day I am so thrilled I found your blog page, I really found you by accident, while I was looking on Aol for something else, Anyways I am here now and would just like to say thanks a lot for a remarkable post and a all round exciting blog (I also love the theme/design), I don’t have time to read it all at the moment but I have bookmarked it and also added in your RSS feeds, so when I have time I will be back to read a great deal more, Please do keep up the excellent work.
Margret
Nice blog here! Additionally your website lots up very fast! What web host are you using? Can I get your affiliate link for your host? I wish my site loaded up as fast as yours lol
Margart
Good answer back in return of this issue with firm arguments and describing all regarding that.
Alica
I love what you guys are usually up too. This sort of clever work and exposure! Keep up the terrific works guys I’ve incorporated you guys to my own blogroll.
Devushki
Нацените красивых девушек на сайте: https://t.me/krasivie_russkie_devushki Спасибо за внимание :)
Dorie
I’m gone to tell my little brother, that he should also pay a quick visit this web site on regular basis to take updated from newest information.
Dinah
Have you ever thought about adding a little bit more than just your articles? I mean, what you say is important and all. But think of if you added some great visuals or videos to give your posts more, “pop”! Your content is excellent but with images and video clips, this site could definitely be one of the very best in its niche.
Awesome blog!
Rolando
Attractive element of content. I simply stumbled upon your weblog and in accession capital to say that I get actually loved account your weblog posts. Any way I will be subscribing for your feeds and even I success you get right of entry to constantly quickly.
Oliver
This is really fascinating, You are an overly professional blogger. I have joined your rss feed and stay up for in quest of more of your fantastic post. Also, I have shared your website in my social networks
Celesta
Valuable information. Lucky me I found your site accidentally, and I am shocked why this twist of fate didn’t came about in advance! I bookmarked it.
Stephania
Hi, its nice paragraph on the topic of media print, we all be aware of media is a enormous source of facts.
Irish
Hello, I want to subscribe for this website to get most recent updates, so where can i do it please help.
Aliza
My partner and I stumbled over here from a different web address and thought I may as well check things out. I like what I see so now i am following you. Look forward to checking out your web page for a second time.
Martina
Spearmint oils can help with pressure reliever.
Implement a tiny amount of essential oil in your temples and the neck and throat if you believe stressed out.
Aliza
Have you ever earned $765 just within 5 minutes? trade binary options
Bernadette
You’ll find delicious vegetarian meals with the help of the VegOut app. Now you may set up the ability supply in the case if it isn’t already put in. The Nook Tablet has not solely a energy button but additionally buttons for volume control. The Kindle Fire has solely a power button. The Kindle Fire and the Nook Tablet every have a dual-core, 1-gigahertz processor. From a hardware perspective, the Nook has the sting over the Kindle Fire. But hardware isn’t the entire story. Or, you’ll be able to rent out the entire property to, say, tourists who need to go to New Orleans however don’t desire to stay in a hotel. Finding just the fitting restaurant is one thing, but getting a reservation might be a complete other headache. These slot machines normally use fruit symbols and you can run the slot machine by clicking on the arm of the machine. Both fashions have an SD-card slot that accepts cards with up to 32 further gigabytes of storage space.
Emanuel
Hey there, You’ve done a great job. I will certainly digg it and personally recommend to my friends. I am sure they will be benefited from this site.
Samantaauj
Приветствую Вас товарищи[url=https://ccwater.kiev.ua/].[/url] http://lat-puzz.com/phpbb/memberlist.php?mode=viewprofile&u=24652 http://galeevmm.ru/forum/memberlist.php?mode=viewprofile&u=373335 http://geartalk.org/forum/viewtopic.php?f=32&t=47823 http://chernigov.info/member.php?u=612512 http://galacticarmada.com/forum/viewtopic.php?f=7&t=65649
Доставка воды в Киеве. По нормам ВОЗ взрослому человеку необходимо не менее 1,5 л чистой воды ежедневно. При этом качество питьевой жидкости не менее важно, чем количество. Оптимальным вариантом считается очищенная столовая вода, предназначенная для ежедневного применения.Самая идеальная вода для утоления жажды, приготовления напитков и блюд для взрослых и детей. Нашу воду в любых объемах можно заказать с доставкой по Киеву в офис или на дом. Самая быстрая доставка питьевой воды в Киеве Ключевым преимуществом компании является быстрая доставка. Уже спустя 60 минут после подтверждения заказа на сайте, вода в указанных объемах прибудет в любую точку Киева. Мы гордимся оперативной и слаженной работой наших сотрудников, которые сделали возможным настолько быструю обработку заказов в условиях мегаполиса. Если по техническим причинам заказ прибыл позже, то вы гарантированно получаете скидку 10% за каждые 10 минут простоя. Заказать воду можно 7 дней в неделю с 8:00 до 20:00, в воскресенье – с 9:00 до 18:00. Операторы колл-центра обрабатывают заказы максимально быстро, а кроме того они дают подробную консультацию по всем вопросам доставки воды, в том числе в оптовых количествах. Источники питьевой воды Наша вода добывается из глубинных скважин на уровне 335 м. Добытая из артезианских источников она проходит тестирование в лаборатории, где определяется минеральный состав и соответствие международным нормам. Многоступенчатая система фильтрации позволяет получить кристально чистую жидкость, идеально сбалансированную по микроэлементному составу воду. Обратите внимание на преимущества нашей продукции: Безопасность – благодаря природным источникам и глубокой очистке наша питьевая вода подходит для детей и взрослых. Экологичность – на глубине бурения наших скважин находятся подземные источники, в которых нет токсинов и вредных химических примесей антропогенного происхождения. Природный состав – вследствие фильтрации жидкость идеально балансируется по микроэлементному составу. Естественный вкус – низкая концентрация минералов позволяет получить воду нейтрального освежающего вкуса, что лучше всего подходит для питья в натуральном виде и приготовления еды. Уже готовую для употребления воду разливают в прочные бутыли, которые после использования могут быть подвержены вторичной обработке, что экологично и современно. Контроль качества питьевой воды Заботясь о своих клиентах, мы гарантируем высокое качество воды. Специалисты компании проверяют ее состав на соответствие международному стандарту FSSC 22000. Помимо регулярного контроля качества питьевой воды, мы проверяем чистоту бутылей, прошедших процедуру автоматической мойки и обеззараживания. Именно поэтому можно утверждать, что питьевая вода принесет только пользу вашему организму. От всей души Вам всех благ!
Hilton
What’s up it’s me, I am also visiting this web site on a regular basis, this web page is actually nice and the users are truly sharing pleasant thoughts.
Stormy
Solar power panels are an easy to set up and might collect sun light for energy. There are many things that needs to be regarded as prior to installing them. The biggest factor you should take into account is just how a lot sun does your house get on average?
Brentsow
эротика порно ролики смотреть бесплатно https://dojki24.club/ смотреть лучшее порно онлайн большие [url=https://dojki24.club/]дойки[/url] смотреть порно натуральные дойки
[img]https://dojki24.club/picture/Milashke-khochet-poprobovat-spermu-na-vkus-i-zabotlivyi-svodnyi-bratets-pomogaet-ei-s-etim.jpg[/img]
[url=http://ljhebblethwaiteltd.co.uk/?cf_er=_cf_process_6334b7e0e0c8b]лучшие порно ролики hd[/url] [url=http://superpottymommy.com/serenity-hassr-taken-posseson/#comment-38340]порно ролики волосатые пизды[/url] [url=https://www.scuolayoga.it/cosa-e-yoga/yoga-a-genova/#comment-59421]порно кончают лучшее онлайн бесплатно[/url] [url=https://yoomoney.ru/transfer/quickpay?requestId=353039393130353035385f31393630353065373531376361633664343063616164633534643764616436643464656163393062]порно большие дойки в автобусе[/url] [url=https://donusenkobi.com/different-post#comment-24408]лучшие порно сайты онлайн бесплатно[/url] [url=http://www.epodismo.com/comments/comments.php]порно хороший кончит рот[/url] [url=http://24tov.com.ua/forum/viewtopic.php?f=12&t=49079&p=249475#p249475]секс порно ролики смотреть онлайн[/url] [url=http://gvsdxpj.webpin.com/?gb=1#top]порно ролики новинки 2021[/url] [url=https://bahamaeats.com/volute-nemo-ipsam-turpis-quaerat-sodales-sapien-2/#comment-17024]скачать хорошее порно видео без регистрации[/url] [url=https://at.alisrv.com/how-does-plastic-end-up-in-the-ocean/#comment-8999]скачать порно дойки без регистрации[/url] 5606153
Jewell
Hey there! Quick question that’s totally off topic.
Do you know how to make your site mobile friendly? My weblog looks weird when browsing from my iphone. I’m trying to find a template or plugin that might be able to correct this issue. If you have any suggestions, please share. With thanks!
Sam
Excellent post! We are linking to this particularly great content on our site.
Keep up the good writing.
Ethel
Hi there! I’m at work browsing your blog from my new iphone! Just wanted to say I love reading through your blog and look forward to all your posts! Keep up the fantastic work!
Ezekiel
I got this web page from my buddy who told me about this web site and now this time I am browsing this website and reading very informative posts at this time.
Trena
Hey there just wanted to give you a quick heads up. The text in your post seem to be running off the screen in Ie. I’m not sure if this is a format issue or something to do with internet browser compatibility but I figured I’d post to let you know. The style and design look great though! Hope you get the problem resolved soon. Kudos
Gary
Its like you read my mind! You seem to know so much about this, like you wrote the book in it or something. I think that you can do with a few pics to drive the message home a little bit, but other than that, this is excellent blog. A fantastic read. I’ll definitely be back.
Tiffany
Hello there! I know this is kinda off topic but I was wondering which blog platform are you using for this website? I’m getting tired of Wordpress because I’ve had issues with hackers and I’m looking at options for another platform.
I would be fantastic if you could point me in the direction of a good platform.
Lizzie
You actually make it seem so easy with your presentation but I find this topic to be actually something that I think I would never understand. It seems too complicated and very broad for me. I’m looking forward for your next post, I will try to get the hang of it!
Roscoe
Right here is the right website for everyone who would like to find out about this topic. You know a whole lot its almost hard to argue with you (not that I personally would want to…HaHa). You definitely put a fresh spin on a topic that’s been discussed for years. Wonderful stuff, just great!
Sol
I don’t even know how I ended up here, but I thought this post was good. I don’t know who you are but certainly you are going to a famous blogger if you are not already ;) Cheers!
Rashad
Aw, this was an exceptionally nice post. Finding the time and actual effort to produce a superb article… but what can I say… I put things off a lot and don’t seem to get anything done.
Charis
Hmm is anyone else encountering problems with the images on this blog loading? I’m trying to find out if its a problem on my end or if it’s the blog. Any suggestions would be greatly appreciated.
Modesto
My brother recommended I may like this blog. He was totally right. This put up truly made my day. You cann’t imagine simply how much time I had spent for this info! Thanks!
Penney
Hello There. I discovered your weblog the usage of msn. This is a very well written article. I will make sure to bookmark it and return to learn more of your useful info. Thank you for the post. I will definitely return.
Carl
If some one wants to be updated with latest technologies therefore he must be pay a quick visit this site and be up to date every day.
Adelaide
Yes! Finally someone writes about frozen shoulder exercises.
Arlette
I blog frequently and I truly appreciate your information. This great article has really peaked my interest.
I will take a note of your blog and keep checking for new details about once per week.
I subscribed to your RSS feed as well.
Tammara
lloyds chemist online viagra where to buy viagra cialis online buy viagra cheaply overnight shipping generic viagra viagra tablet online purchase in india
Rafaela
That laptop computer-ish trait means you will have to look a bit more durable for Internet entry when you are out and about, but you will not should pay a hefty month-to-month fee for 3G knowledge plans.
However the iPad and all of its non-Apple tablet competitors are certainly not an all-encompassing know-how for anyone who wants critical computing energy. It expands on the thought of what pill computer systems are purported to do. This screen also offers haptic feedback in the form of vibrations, which offer you tactile confirmation that the tablet is receiving your finger presses. Because human flesh (and thus, a finger) is a conductor, the screen can precisely decide the place you are pressing and perceive the commands you are inputting. That straightforward USB port additionally may allow you to attach, say, an exterior hard drive, that means you possibly can shortly entry or again up just about any type of content, from photos to text, using the included File Manager app. And for certain forms of games, reminiscent of driving simulators, you’ll be able to turn the pill back and forth like a steering wheel to information movements within the sport. Like its back cowl, the Thrive’s battery can be replaceable. Not only is this helpful if you may be far from a power supply for lengthy periods, nevertheless it additionally permits you to substitute a new battery for one which goes dangerous without having to seek the advice of the manufacturer.
Hilton
Фильмы музыка сериалы онлайн
MatthewMex
How to [url=http://77pro.org/psd-repair.html]repair Photoshop[/url] file.
Kevinbub
ничего подобного
_________
tipobet bedava bonus, [url=https://tr.playrealtopmoneygame.xyz/]iddaa dünkü bugünkü maç sonuçları [/url] - tipobet nasil bir site
Phyllis
paper writing services legitimate websites that write papers for you online paper writing services
Zora
I read this piece of writing fully concerning the difference of hottest and previous technologies, it’s amazing article.
Carin
I think that what you said made a lot of sense. However, think about this, suppose you added a little information? I am not saying your information isn’t solid, however what if you added something that makes people desire more? I mean Heart Disease Demo: How much risk for a single patient has - Dossiers is kinda plain. You should look at Yahoo’s home page and see how they create article titles to get viewers to click. You might try adding a video or a picture or two to grab people interested about everything’ve got to say. In my opinion, it would make your posts a little livelier.
Chasity
Can I just say what a relief to discover an individual who genuinely understands what they are talking about on the web.
You certainly know how to bring an issue to light and make it important. A lot more people need to check this out and understand this side of your story. It’s surprising you’re not more popular since you definitely have the gift.
Arron
Hey! This is kind of off topic but I need some advice from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty quick. I’m thinking about creating my own but I’m not sure where to start. Do you have any points or suggestions? Many thanks
Lara
This design is wicked! You certainly know how to keep a reader amused. Between your wit and your videos, I was almost moved to start my own blog (well, almost…HaHa!) Fantastic job.
I really enjoyed what you had to say, and more than that, how you presented it. Too cool!
Adrianna
дракон в поиске дома смотреть аниме Дом Дракона Сериал Скачать Бесплатно
Tania
great put up, very informative. I wonder why the opposite specialists of this sector do not understand this. You must continue your writing. I’m sure, you have a huge readers’ base already!
Makayla
Thank you, I’ve recently been looking for info about this topic for ages and yours is the best I’ve found out till now. But, what concerning the bottom line? Are you sure about the supply?
Barry
Unquestionably imagine that that you said. Your favorite justification seemed to be on the internet the easiest factor to be mindful of. I say to you, I definitely get annoyed while people think about concerns that they just do not recognize about. You managed to hit the nail upon the top as smartly as defined out the entire thing without having side-effects , other folks can take a signal. Will probably be again to get more. Thank you
ThomasRhino
интересный пост
_________ kapperdən pulsuz futbol proqnozu, [url=https://az.playrealtopmoneygame.xyz/99.html]casino club la rioja[/url], bukmeker kontekstlərini necə təxmin etmək olar
RichardAcutt
Heart Disease Demo: How much risk for a single patient has - Dossiers
Moderator - “:
Лучший сайт для покупки и продажи товаров - МЕГА https://mega-info.xyz . Сегодня MEGA даркнет является самой крупной и известной торговой анонимной площадкой в РФ. Она предлагает свои пользователям доступ к большой базе продавцов из разных стран. Здесь можно приобрести любые товары. Также вы можете сами начать продажу, зарегистрировавшись на проекте. При этом важно понимать, что сайт гарантирует безопасность, а потому проверяет каждого своего продавца. Для этого используются различные способы, в числе которых - тайные покупатели. Потому вы можете быть уверены в качестве покупаемых товаров, честности продавцов, и безопасности покупок на МЕГА онион. А для перехода на сайт, просто использовать активную ссылку MEGA официальный сайт - https://mega-infos.xyz.
[url=https://xealme.com/product/buy-google-reviews/#comment-35146]мега площадка[/url] [url=https://adnmobile.myxwiki.org/xwiki/bin/view/Main/Dashboard/]мега darknet market[/url] [url=https://www.php-quelle.de/erotik-portal-id-251.html]площадка мега[/url] [url=https://journalnews.in/news/newdetail/OTI5]mega магазин[/url] [url=https://pycode.com.br/paste/6nqtk]мега даркнет[/url] 63f2_c3
Felicia
whoah this weblog is excellent i love reading your posts.
Stay up the good work! You realize, lots of persons are looking round for this info, you can help them greatly.
Jeana
Aw, this was an extremely nice post. Spending some time and actual effort to make a really good article… but what can I say… I hesitate a lot and don’t manage to get nearly anything done.
Tangela
Hello i am kavin, its my first time to commenting anyplace, when i read this piece of writing i thought i could also make comment due to this good post.
Tabitha
I pay a quick visit everyday a few web pages and websites to read posts, however this blog provides feature based writing.
Alicia
I pay a quick visit day-to-day a few sites and information sites to read content, but this blog provides feature based content.
Doretha
Hmm it appears like your blog ate my first comment (it was super long) so I guess I’ll just sum it up what I submitted and say, I’m thoroughly enjoying your blog. I as well am an aspiring blog blogger but I’m still new to the whole thing. Do you have any recommendations for first-time blog writers?
I’d really appreciate it.
Pauline
Great blog here! Also your web site a lot up very fast! What web host are you the use of? Can I am getting your associate link in your host? I wish my site loaded up as fast as yours lol
Tyrone
cialis where to buy philippines cialis online dubai generic cialis online pharmacy buy female cialis online cheap cialis 5mg best price
Zachery
nombre generico viagra buy viagra online czech republic natural viagra alternatives generic viagra drugs com viagra tablets online purchase in pakistan
Quentin
where can you buy resume paper online paper writers write my math paper
BitCasino
Bitcoin casinos are online casinos that allow you to deposit, wager and cashout, while using Bitcoin as the currency - sounds simple, right? https://about.me/bitcoin_casino_usa/ Discover the best Bitcoin casinos
Mohammed
It might be difficult to determine which Dragon Ball Super Super Hero update gaming console suits your preferences. Take a look at critiques of several Dragon Ball Super Super Hero movie systems that other avid gamers have identified problems with the gaming console.
BitCasino
Bitcoin casinos are online casinos that allow you to deposit, wager and cashout, while using Bitcoin as the currency - sounds simple, right? https://about.me/bitcoin_casino_usa/ Discover the best Bitcoin casinos
Dell
It stars Halle Berry, Patrick Wilson, John Bradley, Charlie Plummer, Michael Peña, and Donald Sutherland.
ThomasKen
Breaking news, sport, TV, radio and a whole lot more. The [url=https://www.topworldnewstoday.com/]Top World News Today[/url] informs, educates and entertains - wherever you are, whatever your age
Crystle
Hello, Neat post. There’s an issue together with your website in web explorer, might test this? IE nonetheless is the market leader and a large element of other folks will omit your great writing because of this problem.
Marcelino
For newest news you have to visit the web and on web I found this site as a finest web page for hottest updates.
Augusta
Link exchange is nothing else except it is only placing the other person’s weblog link on your page at proper place and other person will also do similar in favor of you.
Jeffry
Thanks for your marvelous posting! I actually enjoyed reading it, you can be a great author. I will ensure that I bookmark your blog and may come back in the foreseeable future.
I want to encourage that you continue your great job, have a nice morning!
Vanita
Hello, I think your site might be having browser compatibility issues. When I look at your blog site in Opera, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, superb blog!
Samantha
I was more than happy to uncover this web site. I wanted to thank you for your time just for this wonderful read!! I definitely really liked every part of it and I have you book-marked to look at new things on your blog.
Eartha
Hi there, I enjoy reading through your post. I like to write a little comment to support you.
Francesco
Hey there would you mind letting me know which web host you’re utilizing? I’ve loaded your blog in 3 completely different browsers and I must say this blog loads a lot quicker then most. Can you suggest a good hosting provider at a honest price? Many thanks, I appreciate it!
Janeen
buy a paper for college buying papers for college can someone write my paper for me
Sanford
paper writing services best write my nursing paper custom written papers
CasinoSajt
Все о казино играх в нашем Телеграм канале - https://t.me/sajt_casino . Благодараю за внимание. Всем хорошей игры :)
BitCasino
Play up to 5000+ online casino games with Bitcoin, Crypto or FIAT currencies ($ € ?) at https://about.me/bitcoin_casino_usa Compare the best Bitcoin casinos in October 2022 - bonuses, rollover, software, and games
massespaf
Приглашаем прийти информацию на тему [url=https://masterokon66.ru/prajs]ремонт окон[/url] если зимой из окон дует . Адрес: masterokon66.ru .
Mahalia
It may also stream simultaneously on paid tiers of Peacock for 60 days.
Davidrip
Breaking news, sport, TV, radio and a whole lot more. The Top [url=https://www.topworldnewstoday.com/]World News Today[/url] informs, educates and entertains - wherever you are, whatever your age
RonaldKal
сочно хорошая порнуха https://pornushnik.vip/ порно видео онлайн порнуха в hd [url=https://pornushnik.vip/]порно[/url] красивая порнуха hd качества
[img]https://pornushnik.vip/picture/Siskastaia-chernokozhaia-zaznoba-vozbudila-belogo-samtsa-dlia-perepikhona.jpg[/img]
[url=http://violetosbourne.co.uk/?page_id=360#comment-30739]порнуха милфы жмж в hd[/url] [url=https://unzippedtv.com/benefits-of-taking-online-classes-and-necessary-help/#comment-9922]порнуха сочные женщины русские[/url] [url=https://www.booksonfire.de/2020/10/18/5-historische-frauen-serie/#comment-12458]лучшая порнуха в чулках hd[/url] [url=https://instantgoldrefining.com/is-it-better-to-buy-gold-bars-or-gold-coins/#comment-873]порнуха 2021 hd качестве[/url] [url=http://www.meublesdeco.be/vanavond-eten-we-puur-oosters/#comment-29374]бесплатное видео hd порнуха[/url] [url=https://grankom.com.ua/market/schetchiki-elektroenergii/ace-6000-100a.html]перевод на русском языке hd ххх порнуха[/url] [url=http://www.transiberiana.ru/2011/09/12/litexp-project/#comment-179806]лучшая порнуха full hd[/url] [url=http://fly.historicwings.com/policies-and-rights/comment-page-1/#comment-1345908]порнуха секс брызги спермы hd[/url] [url=https://zabor164.ru/components/com_chronoforms/chrono_verification.php?imtype=0]порнуха мать hd[/url] [url=http://avventa.electrob.ru/board/page_0/Stanciya_progreva_betona_SPBM/19251/]день порнуха в hd[/url] 000c856
Dulcie
I like what you guys are up too. This sort of clever work and coverage! Keep up the amazing works guys I’ve included you guys to our blogroll.
Josefina
Hmm it appears like your blog ate my first comment (it was extremely long) so I guess I’ll just sum it up what I wrote and say, I’m thoroughly enjoying your blog. I as well am an aspiring blog blogger but I’m still new to everything. Do you have any recommendations for rookie blog writers? I’d definitely appreciate it.
Homer
Great beat ! I would like to apprentice while you amend your web site, how can i subscribe for a blog site? The account aided me a acceptable deal. I had been tiny bit acquainted of this your broadcast offered bright clear idea
Harley
Hi, i think that i saw you visited my site so i came to go back the choose?.I am attempting to in finding things to enhance my website!I suppose its ok to make use of a few of your ideas!!
Eugenia
Hello, everything is going well here and ofcourse every one is sharing facts, that’s in fact good, keep up writing.
Rene
Everyone loves what you guys tend to be up too. This sort of clever work and reporting! Keep up the great works guys I’ve included you guys to my personal blogroll.
Maggie
Simply wish to say your article is as surprising. The clarity in your post is just great and i could assume you are an expert on this subject. Well with your permission let me to grab your feed to keep up to date with forthcoming post. Thanks a million and please continue the gratifying work.
Letha
Hey There. I found your blog using msn. This is a very well written article. I will be sure to bookmark it and return to read more of your useful information. Thanks for the post. I will certainly comeback.
Aleisha
buy daily cialis cialis 20 mg online kopen cialis 5mg best price legit place to buy cialis online 5mg cialis online
Brayden
As if the tulips in bloom, aren´t enough reason already to go to Amsterdam in spring.
Crystle
After touching one, she experiences surreal hallucinations earlier than waking up again home later that evening.
Lamar
college papers to buy write my economics paper buying college papers
Leautheeraccut
[url=http://77pro.org/27-ost-viewer-03.html]OST Viewer[/url]
Krystyna
Hi, i think that i saw you visited my weblog thus i came to “return the favor”.I’m attempting to find things to enhance my website!I suppose its ok to use some of your ideas!!
Charissa
Wonderful blog! Do you have any tips for aspiring writers?
I’m hoping to start my own website soon but I’m a little lost on everything. Would you propose starting with a free platform like Wordpress or go for a paid option? There are so many options out there that I’m completely overwhelmed .. Any suggestions? Kudos!
Robbin
Very good article! We are linking to this particularly great article on our site. Keep up the good writing.
Annabelle
Hi! Would you mind if I share your blog with my myspace group? There’s a lot of folks that I think would really enjoy your content. Please let me know. Thanks
Pedro
Hi friends, good piece of writing and nice arguments commented at this place, I am genuinely enjoying by these.
Yetta
You can definitely see your expertise within the work you write. The arena hopes for more passionate writers like you who are not afraid to say how they believe. Always follow your heart.
Chanel
This site was… how do you say it? Relevant!!
Finally I have found something which helped me. Kudos!
Antonio
In April, James Cusati-Moyer, Bodhi Sabongui, Mo Amer, and Uli Latukefu joined the solid in undisclosed roles.
Loreen
I’d like to find out more? I’d care to find out more details.
Barry
Greetings! I know this is kinda off topic nevertheless I’d figured I’d ask. Would you be interested in trading links or maybe guest writing a blog article or vice-versa? My site goes over a lot of the same subjects as yours and I believe we could greatly benefit from each other. If you are interested feel free to shoot me an email. I look forward to hearing from you! Excellent blog by the way!
Kimberly
This excellent website truly has all the info I needed concerning this subject and didn’t know who to ask.
Reed
Thank you a lot for sharing this with all folks you really understand what you are speaking approximately! Bookmarked. Please also seek advice from my website =).
We will have a link alternate arrangement between us
Mariam
You’ve made some really good points there. I looked on the net for more information about the issue and found most people will go along with your views on this site.
Corina
I was suggested this website by my cousin. I am not sure whether this post is written by him as no one else know such detailed about my difficulty. You’re incredible! Thanks!
Tobias
Hi there to every body, it’s my first go to see of this web site; this webpage contains remarkable and genuinely excellent material in support of readers.
Bryce
Hi there, its pleasant article about media print, we all understand media is a enormous source of facts.
Larryreets
Ростовский центр юридической помощи ООО “Закон и порядок” предлагает услуги: [url=https://xn—-7sbcfb9bujbbasekfjbg.xn–p1acf/]о списании долгов по кредитам[/url]
Бесплатные консультации, расчет рисков и последствий банкротства.
Huey
If three castles are available in view, Lucky Count awards you 15 free spins. The Lucky Count slot machine comes with five reels and 25 paylines. While most slots games function simply the one wild symbol, Lucky Count comes with two! And regardless of being what CNet calls a “minimalist gadget,” the Polaroid Tablet still has some pretty nifty hardware options you’d anticipate from a extra expensive tablet by Samsung or Asus, and it comes with Google’s new, feature-wealthy Android Ice Cream Sandwich working system. Davies, Chris. “Viza Via Phone and Via Tablet get Official Ahead of Summer Release.” Android Community. You’ll additionally get a free copy of your credit score report – test it and keep in contact with the credit bureaus till they right any fraudulent expenses or accounts you find there. I took this alternative to sign up for the RSS feed or e-newsletter of every one in every of my sources, and to get a copy of a 300-web page government report on power sent to me as a PDF. You’ll additionally get the possibility to land stacks of wilds on an excellent multiplier so this fearsome creature might develop into your best buddy. It boasts a thrilling ride on excessive volatility and is effectively value a spin on VegasSlotsOnline to test it out free of charge.
Aliza
It’s very trouble-free to find out any topic on net as compared to textbooks, as I found this article at this web site.
Margarita
Hi to all, how is the whole thing, I think every one is getting more from this website, and your views are pleasant in support of new people.
Akilah
Hi there, I enjoy reading through your article. I wanted to write a little comment to support you.
Davida
Hi, i think that i saw you visited my web site so i came to “return the favor”.I’m attempting to find things to improve my site!I suppose its ok to use some of your ideas!!
Dian
I visited several web pages but the audio feature for audio songs current at this web site is actually superb.
Jina
Fantastic beat ! I wish to apprentice while you amend your site, how could i subscribe for a weblog site? The account helped me a acceptable deal. I have been a little bit familiar of this your broadcast offered bright clear concept
Charles
Very nice post. I just stumbled upon your weblog and wanted to say that I have truly enjoyed browsing your blog posts. In any case I’ll be subscribing to your rss feed and I hope you write again soon!
Darell
Hi there, yes this piece of writing is genuinely fastidious and I have learned lot of things from it regarding blogging. thanks.
Delphia
I think the admin of this web page is truly working hard in support of his web site, since here every material is quality based material.
Coy
Keep on working, great job!
Tabatha
I loved as much as you’ll receive carried out right here. The sketch is attractive, your authored subject matter stylish. nonetheless, you command get bought an shakiness over that you wish be delivering the following. unwell unquestionably come more formerly again as exactly the same nearly very often inside case you shield this hike.
Dominik
If you are going for finest contents like me, simply go to see this site every day for the reason that it provides feature contents, thanks
Terrie
With havin so much written content do you ever run into any problems of plagorism or copyright violation? My blog has a lot of exclusive content I’ve either created myself or outsourced but it seems a lot of it is popping it up all over the web without my permission. Do you know any techniques to help reduce content from being ripped off? I’d really appreciate it.
Maritza
I will immediately clutch your rss as I can not in finding your email subscription link or e-newsletter service. Do you have any? Please allow me recognize so that I could subscribe. Thanks.
Charlene
I was wondering if you ever considered changing the layout of your website? Its very well written; I love what youve got to say. But maybe you could a little more in the way of content so people could connect with it better. Youve got an awful lot of text for only having 1 or 2 pictures. Maybe you could space it out better?
Maryjo
This text is worth everyone’s attention. Where can I find out more?
Christine
After the VM updates lessons with the brand new variations of fields and features, the Flutter framework mechanically rebuilds the widget tree, permitting you to quickly view the results of your changes. The framework includes Hot-reload.
This template consists of greater than 60 screens with very clean animation that is according to Android and IOS. Example labels of those two duties are proven in the bottom two rows of Table 1. Segment tagging and named entity tagging can be thought to be syntactic labeling, while slot filling is extra like semantic labeling. Just like Tesco, the first accessible delivery slot for ASDA is on the 14th of April, although, the supermarket currently appears to be having trouble with their webpage as the grocery part is currently down. And for good cause: It was slow, it regarded totally different than advertised, there have been no USB ports and not using a bulky adapter, the microSD memory card slot wasn’t spring loaded, so it was practically unattainable to get the card out. There’s a protracted checklist of roles players can select from, and Star Atlas has deliberate out a large in-recreation economy the place customers can mine for a revenue and even secure DeFi loans and insurance coverage insurance policies on ships.
Phoebe
Looking to make a change from the town life to the country life?
EvoToimi
Legal casinos, which are state-run, but still owned by private contractors, are available in Germany https://about.me/superspieler Find the best online casino Germania with bonuses and spins
Thoomaasneich
[url=http://77pro.org/28-pst-viewer-09.html]PST Viewer[/url]
Astopoush
Nivolumab and Ipilimumab Effective against Melanoma That Has Spread to the Brain <a href=http://buylasixon.com/>hctz vs lasix</a>
alosync
The study groups participated in median of three sessions per week, 30 40 min each of moderate exercise and were found to have decreased percent body fat, without significant change in weight or lean body mass <a href=http://buylasixon.com/>lasix not working for edema</a>
Boyd
website that writes papers for you write my paper for me cheap pay to do paper
RiddToimi
Online kasiino saidid erinevad mitmete kriteeriumide poolest, mida tuleb arvesse votta, et valida parim sait: https://steemit.com/kasiino/@kasinoid/mis-on-garantii-online-kasiino-terviklikkuse-kohta Parimad online kasiinod leiad alati Eesti Kasiino lehelt.
Theodore
I’m really inspired together with your writing talents and also with the format on your weblog.
Is this a paid topic or did you modify it your self? Either way stay up the nice quality writing, it’s uncommon to look a nice weblog like this one nowadays..
Candace
custom paper writing services i don’t want to write my paper instant paper writer
Valentinaof4
TikTok star known for dance videos in which she wears colorful party clothes -https://valentinaof4.com/ She has earned more than 20 million followers
Trinidad
Hello everyone, it’s my first pay a quick visit at this web site, and post is really fruitful designed for me, keep up posting such content.
Porter
A motivating discussion is worth comment. I believe that you should write more on this issue, it may not be a taboo subject but typically folks don’t speak about such issues. To the next! Kind regards!!
Edwina
I delight in, cause I found just what I was looking for. You’ve ended my four day lengthy hunt! God Bless you man. Have a nice day. Bye
Darnell
Greetings! This is my 1st comment here so I just wanted to give a quick shout out and tell you I truly enjoy reading your posts. Can you recommend any other blogs/websites/forums that deal with the same subjects? Thank you so much!
Juan
This is my first time visit at here and i am really impressed to read all at one place.
Sarah
Wow, incredible blog format! How long have you been blogging for?
you made running a blog glance easy. The total look of your site is magnificent, let alone the content!
Mose
Very good information. Lucky me I recently found your website by chance (stumbleupon).
I’ve book marked it for later!
Leroy
Saved as a favorite, I like your web site!
Dominick
Magnificent beat ! I wish to apprentice while you amend your website, how could i subscribe for a blog web site? The account helped me a appropriate deal. I had been a little bit acquainted of this your broadcast provided bright clear idea
Filomena
This is my first time visit at here and i am truly impressed to read all at single place.
Clyde
My family always say that I am wasting my time here at web, but I know I am getting familiarity every day by reading thes nice articles or reviews.
Carmel
Most of those sites have a mobile app for watching their movies.
Ulrike
Directed by Shi and produced by Lindsey Collins, Disney and Pixar’s “Turning Red” is now streaming solely on Disney+.
Zella
Cooper talked to UCR in September in regards to the intricacies of his stage show and his pleasure to resume touring after greater than a 12 months off the street because of the coronavirus pandemic. For common diners, it’s a fantastic way to learn about new eateries in your area or discover a restaurant when you’re on the highway. Using the operate of division into classes, you can simply discover something that will suit your style. But DVRs have two major flaws – you must pay for the privilege of utilizing one, and you’re stuck with no matter capabilities the DVR you purchase occurs to come with.
This template is appropriate for any working system, due to this fact, utilizing this template is as easy as booking a hotel room. Therefore, it’s perfectly appropriate for the design of a weblog utility.
Therefore, not solely the furnishings needs to be snug, but also the application for its purchase.
Proximus
Smartproxy is one of the best proxy provider services you can find for residential and business use: https://www.facebook.com/bestproxyservers There are quite a few features that make Smartproxy one of the top contenders in the best proxy server service space
Winnie
It’s going to be ending of mine day, however before end I am reading this impressive post to improve my experience.
Deloras
If three castles come in view, Lucky Count awards you 15 free spins. The Lucky Count slot machine comes with 5 reels and 25 paylines. While most slots video games feature simply the one wild symbol, Lucky Count comes with two! And despite being what CNet calls a “minimalist machine,” the Polaroid Tablet nonetheless has some fairly nifty hardware options you’d anticipate from a more expensive pill by Samsung or Asus, and it comes with Google’s new, characteristic-rich Android Ice Cream Sandwich working system. Davies, Chris. “Viza Via Phone and Via Tablet get Official Ahead of Summer Release.” Android Community. You’ll also get a free copy of your credit score report – examine it and stay in touch with the credit score bureaus until they right any fraudulent charges or accounts you discover there. I took this opportunity to join the RSS feed or e-newsletter of each one among my sources, and to get a duplicate of a 300-web page government report on power sent to me as a PDF. You’ll also get the possibility to land stacks of wilds on a very good multiplier so this fearsome creature may change into your finest buddy. It boasts a thrilling trip on high volatility and is well price a spin on VegasSlotsOnline to check it out without cost.
Polly
Hi, I do believe this is an excellent site. I stumbledupon it ;) I may come back once again since I book marked it. Money and freedom is the greatest way to change, may you be rich and continue to guide other people.
Ladonna
Good day! Would you mind if I share your blog with my myspace group? There’s a lot of people that I think would really appreciate your content. Please let me know. Many thanks
Micaela
Having read this I thought it was really enlightening. I appreciate you spending some time and energy to put this informative article together.
I once again find myself spending a lot of time both reading and posting comments. But so what, it was still worthwhile!
Joycelyn
Thank you a bunch for sharing this with all folks you really understand what you’re talking about! Bookmarked. Please additionally discuss with my web site =). We may have a link trade agreement between us
Eric
Touche. Great arguments. Keep up the great effort.
Robertbog
порно ролики инцест онлайн https://razvodila.xyz/ порно ролики инцеста без [url=https://razvodila.xyz/]порно[/url] порно ролики лишения
[img]https://razvodila.xyz/picture/Zanimaetsia-s-boifrendom-seksom-poka-papa-doma.jpg[/img]
[url=https://us.lexusownersclub.com/videos/lexus-videos/2022-lexus-es-to-be-unveiled-on-april-18-in-shanghai-r1087/page/473/#comment-26945]смотреть порно ролики онлайн в hd[/url] [url=https://orehovo-zuevo.dnk-otcovstvo.ru/]порно ролики зрелые женщины[/url] [url=https://itexam.blog.ss-blog.jp/2016-03-21-15?comment_success=2022-10-10T18:54:39&time=1665395679]порно ролики passion hd скачать бесплатно[/url] [url=http://www.moto.pieniadz.pl/Renault,Clio,RS,200,EDC,i,RS,220,Trophy,przeszly,lifting,1-74467-1.html#kom_last]смотреть фильмы онлайн порно ролики[/url] [url=http://bakar11-11.dk/guestbook/guestbook.php]порно ролики мамки hd[/url] [url=http://onlycracked.com/blog/wp-testimony-plus-settings/#comment-8959]порно ролики hd через торрент[/url] [url=https://favoritour.com/parks-and-carnivals/pure-luxe-in-punta-mita/8012-2018/#comment-241309]порно ролики делают минет[/url] [url=http://aurorahcs.com/forum/viewtopic.php?f=8&t=255642]порно ролики большие девушки[/url] [url=https://f1ms.blog.ss-blog.jp/2018-11-17?comment_success=2022-10-08T23:19:12&time=1665238752]смотреть видео порно ролики hd[/url] [url=https://paynesmonuments.webs.com/apps/guestbook/]порно с родителями ролики hd[/url] 4714b7a
JonkisHD
The Proxy site is the infrastructure built to remove barriers to websites being banned https://telegra.ph/Types-of-proxy-servers-10-10 Using a proxy server is one way to hide ip address, and browse anonymously
Warner
Howdy very nice site!! Man .. Excellent .. Amazing .. I will bookmark your web site and take the feeds also? I’m happy to find numerous useful info right here in the post, we’d like develop more techniques in this regard, thank you for sharing. . . . . .
Ellis
Hello to all, how is the whole thing, I think every one is getting more from this web page, and your views are good designed for new people.
Vania
Good day I am so excited I found your website, I really found you by error, while I was looking on Askjeeve for something else, Anyhow I am here now and would just like to say many thanks for a incredible post and a all round entertaining blog (I also love the theme/design), I don’t have time to read it all at the moment but I have saved it and also added your RSS feeds, so when I have time I will be back to read more, Please do keep up the awesome job.
Marta
Have you ever thought about creating an ebook or guest authoring on other sites? I have a blog centered on the same ideas you discuss and would love to have you share some stories/information. I know my readers would value your work. If you’re even remotely interested, feel free to shoot me an email.
Winnie
Paragraph writing is also a fun, if you be acquainted with after that you can write otherwise it is complex to write.
Roxie
We’re a group of volunteers and starting a new scheme in our community. Your web site offered us with valuable information to work on. You’ve done an impressive job and our entire community will be thankful to you.
Ricardo
I blog frequently and I really appreciate your content. This article has really peaked my interest. I will book mark your blog and keep checking for new details about once a week. I subscribed to your Feed too.
Janine
I was wondering if you ever thought of changing the structure of your site? Its very well written; I love what youve got to say. But maybe you could a little more in the way of content so people could connect with it better. Youve got an awful lot of text for only having one or two images. Maybe you could space it out better?
Gennie
Hi my family member! I want to say that this article is awesome, great written and include approximately all important infos. I would like to peer extra posts like this .
Gena
Revelation Puzzle Rooms also has a room where people can track down the person who created the virus and another, titled Toxic, which challenges players to create a cure for the virus.
RobertSed
порно домашнее анал первый раз https://fakporn.xyz/ индийское домашнее порно [url=https://fakporn.xyz/]порно[/url] смотреть порно домашнее зрелые
[img]https://fakporn.xyz/picture/Russkaia-koketka-Izabella-Klark-podstavila-popku-pod-kher-partnera.jpg[/img]
[url=http://subvert.pw/f/viewtopic.php?pid=109548#p109548]жесткое порно смотреть онлайн качество hd[/url] [url=http://kristenanna.com/index.php/2019/04/11/the-secret-ive-been-keeping/#comment-17530]домашнее порно соло[/url] [url=https://www.communityheritagemaps.com/changing-tides-of-shopping/#comment-52506]домашнее любительское порно студентки[/url] [url=https://luillu.pl/pl/n/6#comment13104]студийное порно анал[/url] [url=https://asked4afriend.com/2020/07/07/hello-world/#comment-6443]смотреть порно домашний попа[/url] [url=https://patisserie-tanger.de/robusta/vogelnest/#comment-1174]любительское жена кончает русское порно[/url] [url=https://justlada.ru/nashi-lyudi/469-vaz-2102.html]домашнее порно с собакой[/url] [url=http://www.h370.com/forum.php?mod=viewthread&tid=2163516&extra=]порно домашнее кончают подборка[/url] [url=http://www.ewaldgarage.nl/2013/12/09/commentaar/#comment-25648]порно секс русское домашнее инцест[/url] [url=https://italynlove.it/2020/05/12/mascherina-fashion-must-have-2020/#comment-237754]смотреть русское домашнее порно зрелых[/url] 163f3_2
Amber
This piece of writing will assist the internet visitors for building up new blog or even a weblog from start to end.
Slava
Дядя Слава открыл рот https://t.me/djadja_slava Современный Дядя Слава, или Дед, как его еще называют, – тоже человек с богатым криминальным прошлым.
etherExink
Займы онлайн в Благовещенске [url=https://coursestop.ru/hplv]Микрозайм онлайн срочно в Электростали мгновенные займы на карту онлайн быстро[/url]
онлайн займ переводом мини займы онлайн круглосуточно на карту срочные займы онлайн с плохой кредитной историей
<a href=https://shoploans.ru/v3s7>взять займ онлайн срочно без отказа займы Пенза</a>
Tory
Hello to all, the contents existing at this web site are in fact amazing for people experience, well, keep up the good work fellows.
Lino
Do you have any video of that? I’d care to find out more details.
Jurius
Online kazino celvedis Latvija. Labakie tiessaistes kazino - spelu automati, bonusi un bezriska griezieni - https://t.me/online_kazino_latvija/4 Visplasakais vietejo un arzemju kazino saraksts.
DwainLauts
Probably, I am mistaken. https://fuckmilf.info https://amazonxpeditions.com
Michaelliels
https://bookmarkmiracle.com/story1670876/5-tips-about-online-kazino-you-can-use-today https://bookmarkmiracle.com/story1670876/5-tips-about-online-kazino-you-can-use-today
Jarrod
We also help our purchasers with financial institution deposit to file citizenship purposes.
Selena
dltk custom writing paper help writing college papers best college paper writing service
Sdvillhob
[url=https://chimmed.ru/products/search?name=%D1%85%D0%BB%D0%BE%D1%80%D0%BE%D1%84%D0%BE%D1%80%D0%BC]хлороформ купить [/url] Tegs: моксидектин https://chimmed.ru/products/moksidektin-id=1511012
[u]сульфаметоксазол [/u] [i]ампициллина тригидрат [/i] [b]муравьиная кислота купить [/b]
Jared
buy papers online for college will you write my paper for me write my english paper
Bnuolmut
Unsurlily [url=https://1xbetbonuses.com/]1xBet Bonus[/url] phototopographic
Tyrell
A key enchancment of the new ranking mechanism is to mirror a extra correct choice pertinent to recognition, pricing policy and slot effect based on exponential decay mannequin for online users. This paper studies how the online music distributor ought to set its ranking coverage to maximize the worth of on-line music ranking service. However, earlier approaches usually ignore constraints between slot worth representation and related slot description representation in the latent space and lack sufficient mannequin robustness. Extensive experiments and analyses on the lightweight fashions present that our proposed methods obtain considerably increased scores and substantially improve the robustness of each intent detection and slot filling. Unlike typical dialog fashions that rely on large, complicated neural community architectures and huge-scale pre-trained Transformers to attain state-of-the-artwork results, our methodology achieves comparable outcomes to BERT and even outperforms its smaller variant DistilBERT on conversational slot extraction duties. Still, even a slight enchancment might be value the fee.
SuperDedushka
Подборка забавных и немного пошлых фотографий с красивыми девушками - https://t.me/i_kakie_devushki красивые девушки спортсменки, сексуальные девушки в спорте, фото и видео
Michaelliels
[url=https://ndt-innovation.ru/]Компания НК-Инновации ЮГ предлагает широкий выбор устройств для диагностики в Москве и Самаре, которые можно использовать в различных промышленных отраслях. В нашем каталоге представлены устройства, позволяющие осуществлять контроль герметичности и твердости, изоляции и среды. Мы предлагаем диагностическое оборудование в Москве и Самаре для разных разрушающих и неразрушающих способов контроля:
акустического; капиллярного; визуального; магнитного; рентгеновского; ультразвукового и других. Это может быть лабораторное оборудование или устройства, используемые в «полях», технические средства для проведения химического анализа, для анализа металлов и сплавов. Мы предлагаем широкий выбор оборудования для диагностики, причем делаем это на выгодных для вас условиях.
Вся предлагаемая нами техника прошла многократные испытания и имеет все необходимые сертификаты. Это действительно надежное диагностическое оборудование, выполняющее диагностику с минимальной погрешностью и обладающее продолжительным сроком службы.[/url]
Alexander
buy college papers ghost writer college papers where can i buy resume paper
NugUttefe
In normal mammary epithelial cells, STAT5A is the dominant actor, whereas in malignant mammary epithelial cells, STAT5B can be the more significant contributor <a href=http://bestcialis20mg.com/>real cialis online</a>
Tutinhent
<a href=http://bestcialis20mg.com/>cialis for sale in usa</a> A functioning city needs to attract some wealthy people who can hire other people and contribute to the tax base
Sheryl
write my paper for me fast help writing a college paper professional college paper writers
Progon
Оптимальным решением в вопросе оптимизации сайта станет прогон хрумером на сайте https://t.me/progon_sajta_hrumerom Максимальный прогон и размещение через программу Xrumer по свежим и актуальным базам.
Michaelliels
[url=https://ndt-innovation.ru/]Компания НК-Инновации ЮГ осуществляет быструю поставку диагностического оборудования предприятиям в Москве, Самаре и в других регионах России. Зачастую от скорости поставки таких устройств зависит стабильность работы нефтяных скважин, производственных предприятий или других объектов. Оперативность поставки диагностического оборудования достигается нами благодаря тесному сотрудничеству со многими российскими и зарубежными производителями. Это же позволяет предлагать устройства для диагностики по доступным ценам.
У нашей компании многолетний опыт работы в сфере поставки технического диагностического оборудования. Мы оперативно поставляем не только саму технику, но и необходимые для работы с ней расходные материалы. В нашей компании работают опытные компетентные специалисты, готовые помочь вам в выборе оборудования и в формировании заказа на его поставку.
Обращаясь в НК-Инновации ЮГ, вы можете рассчитывать на высокое качество диагностического оборудования, низкие цены, профессиональные консультации и сервисное обслуживание. [/url]
Don
After checking out a number of the blog posts on your site, I seriously appreciate your technique of writing a blog. I book marked it to my bookmark site list and will be checking back soon. Please visit my web site as well and let me know how you feel.
Krista
These vary from 2x to 8x for the chance of much greater prizes.
The Komodo Dragon slot machine’s 50 paylines are fastened and you can choose from a spread of wagers beginning from 0.25 to a max wager of 50.00. It’s a decent selection in your common gambler however it probably won’t appeal to the high rollers. The road wins are multiplied with the credits you choose to wager per line. You choose the variety of lines and bet per line which make up the whole win. It’s a fearsome ally as it lands in stacks for further probabilities at multiple win strains. It’s possible to score a full matrix of dragon wilds for bumper prizes. Wilds can act as others if they can then full a win, and with enough in view, you possibly can simply acquire multiple prizes on the last spin in a collection. Fans and producers labored exhausting to save the sci-fi sequence “Quantum Leap” from the notoriously dangerous eight p.m. You’ll additionally get the possibility to land stacks of wilds on a great multiplier so this fearsome creature may change into your best good friend.
Wallace
will you write my paper for me online paper writers write my english paper for me
Yvette
Amazing things here. I’m very satisfied to look your post. Thank you a lot and I’m taking a look ahead to touch you.
Will you kindly drop me a mail?
Anja
I was able to find good info from your articles.
Wilbert
Hello there! Do you know if they make any plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any suggestions?
Louanne
Hi there Dear, are you actually visiting this site on a regular basis, if so then you will without doubt get nice know-how.
Antonetta
I got this website from my friend who told me regarding this web site and now this time I am visiting this website and reading very informative posts at this time.
Ellis
Link exchange is nothing else except it is only placing the other person’s webpage link on your page at appropriate place and other person will also do similar in favor of you.
Violette
You could certainly see your skills in the article you write. The world hopes for even more passionate writers like you who are not afraid to say how they believe. At all times follow your heart.
Cristine
Hi there to every one, the contents existing at this site are really amazing for people experience, well, keep up the nice work fellows.
Jamesinoke
[url=http://77pro.org/47-08-beschaedigten-powerpoint-datei-machen.html]repair[/url]
Xrumer
Как правильно сделать прогон Хрумером узнайте на канале https://t.me/progon_sajta_hrumerom/426 Для того чтобы ХРумер был эффективен, необходимо как следует разобраться во всех тонкостях работы с этой программой
Williamspake
порно секс онлайн телефона https://pizdasja.top/ порно инцесты снятые на телефон [url=https://pizdasja.top/]порно[/url] мобильный бесплатное домашнее порно
[img]https://pizdasja.top/picture/Rasputnaia-blondinka-Andi-Anderson-gotova-poluchit-penisy-v-svoi-dyrki.jpg[/img]
[url=https://anzai-mfg.com/publics/index/68/b_id=424/r_id=2/fid=d890636882398a0c6fcbb24cdb68dc20]мобильное порно мать сын[/url] [url=http://cgi.members.interq.or.jp/venus/awabi/awabijinja/bbs08/index.html]секс порно видео женой бесплатно[/url] [url=http://usgkoqj.webpin.com/?gb=1#top]порно видео пока разговаривает по телефону[/url] [url=http://shop.aviva-telecom.ru/blog/pryamye-postavki-s-kitaya#comment_20604]ххх реал домашний секс[/url] [url=https://elexbetcasino.com/2020/08/26/elexbet-hakkinda/#comment-8204]девочке секс порно ххх[/url] [url=http://voindom.ru/flat/mobilnoe-porno-vtroem]мобильное порно втроем[/url] [url=http://www.sahnehaeubchen.org/#comment-29714]бесплатное порно в чулках на телефон[/url] [url=http://babsib.at/cms/wb/pages/gaestebuch.php]скачать телефон порно страпон[/url] [url=https://www.theagiler.com/dipendente-lavoratore-ma-dipendente-da-cosa/#comment-154867]домашнее порно видео снятое на телефон[/url] [url=https://ubrukopi.com/dev/product/timemore-dripper-crystal-eye-02-acrylic/#comment-21985]русское порно с мобильного телефона с разговорами[/url] 5_4878c
KizSkali
A lot of thanks for all your hard work on this blog and alo try to [url=https://www.kizi10.org] play kizi10 games [/url]. Ellie takes pleasure in setting aside time for investigations and it’s really simple to grasp why. My spouse and i hear all concerning the compelling method you deliver powerful items on this website and as well strongly encourage participation from website visitors on the area plus my princess is undoubtedly being taught so much.
Deangelo
I know this site offers quality based articles and additional material, is there any other web site which presents these stuff in quality?
Leonel
These are actually fantastic ideas in about blogging. You have touched some nice points here. Any way keep up wrinting.
Dinah
Wow, fantastic blog layout! How lengthy have you ever been running a blog for? you make blogging glance easy. The overall look of your website is magnificent, let alone the content material!
Derrick
You are so interesting! I do not think I have read through anything like that before. So wonderful to discover someone with a few unique thoughts on this topic. Seriously.. thanks for starting this up. This site is one thing that is required on the web, someone with a bit of originality!
Cornell
What’s up, this weekend is nice in support of me, since this moment i am reading this wonderful informative piece of writing here at my residence.
Hortense
Hello There. I found your blog using msn. This is a really well written article. I will make sure to bookmark it and come back to read more of your useful information. Thanks for the post. I’ll certainly return.
Elke
Hello, Neat post. There’s a problem along with your site in web explorer, could test this? IE nonetheless is the market chief and a good section of other folks will pass over your fantastic writing due to this problem.
Maryanne
I’ve been browsing online greater than 3 hours nowadays, but I by no means discovered any fascinating article like yours. It’s lovely value enough for me. In my opinion, if all web owners and bloggers made just right content material as you did, the net will be a lot more helpful than ever before.
Les
custom college paper will someone write my paper for me write my paper in 3 hours
PinUp
Рейтинг онлайн казино включает список лучших виртуальных игорных клубов с возможность играть в игровые автоматы, рулетку, блэкджек и другие азартные игры и конечно вход в казино “Зеркало” через Телеграм канал https://t.me/pin_up_casino_vhod_zerkalo . Топ лучших онлайн казино на реальные деньги.
Gretchen
отчаянные домохозяйки смотреть онлайн
Otilia
My family members always say that I am killing my time here at net, except I know I am getting know-how daily by reading thes nice content.
Celia
мультик канителька капризулька
Kumarhane
Kumarhane.com’un onerileri - https://t.me/kumarhane_online ilmege deger hususlar Her ne kadar internet gazinolar Amerikan isletmecilerin elinde ise de bunlar?n isyerleri Amerika’da degil, bilakis cogu Karayip bolgesinde
Shantejeday
Хочу купить
[url=https://t.me/HiFiport/3013]XDuoo[/url]. Какие посоветуете, ребят?
Progonov
Добрый день а сайт на блогеер можете прогнать? https://telegra.ph/5-prostyh-shagov-dlya-provedeniya-otlichnoj-SEO-kampanii-10-17 Прогон можно делать и молодому сайту. Ссылки индексируются в течении 1,5-2 месяцев, постепенно, что никак отрицательно не влияет на сайт.
Virgie
This article will assist the internet people for creating new weblog or even a blog from start to end.
Silas
Hello there, just became alert to your blog through Google, and found that it is really informative. I am gonna watch out for brussels. I’ll be grateful if you continue this in future. Many people will be benefited from your writing. Cheers!
Candy
Its such as you read my thoughts! You appear to understand so much approximately this, such as you wrote the ebook in it or something. I believe that you simply could do with some percent to force the message home a little bit, but other than that, this is wonderful blog. An excellent read. I will definitely be back.
Hans
Thank you for the auspicious writeup. It in fact was a amusement account it. Glance complex to far introduced agreeable from you! By the way, how can we communicate?
Laurinda
This is a topic that is near to my heart…
Thank you! Exactly where are your contact details though?
Fabian
If you would like to increase your knowledge simply keep visiting this web site and be updated with the most recent gossip posted here.
Phil
If some one desires to be updated with latest technologies after that he must be go to see this website and be up to date every day.
Eli
Hi! I know this is somewhat off topic but I was wondering if you knew where I could get a captcha plugin for my comment form?
I’m using the same blog platform as yours and I’m having trouble finding one?
Thanks a lot!
Angelika
I appreciate, result in I found exactly what I used to be having a look for. You’ve ended my four day lengthy hunt! God Bless you man. Have a great day. Bye
RonaldBon
порно фото женщины скачать на телефон https://kisuni.xyz/ скачать бесплатно порно на телефон 2 [url=https://kisuni.xyz/]порно[/url] скачать порно проституток бесплатно на телефон
[img]https://kisuni.xyz/picture/Aziatka-trakhaet-paltsami-i-vibratorom-svoe-zarosshee-vlagalishche.jpg[/img]
[url=https://castletheatremanchester.org/#comment-32272]порно видео скачать сосет на телефон[/url] [url=http://gezirasocial.gov.sd/?p=984#comment-1788942]украинское порно скачать телефон[/url] [url=https://k53u-sxe450.blog.ss-blog.jp/2016-07-27-1?comment_success=2022-10-14T16:14:34&time=1665731674]порно скачать на телефон масло[/url] [url=http://probeauty.com.ru/blog/alfaparf-#comment_119656]голые девушки порно скачать на телефон[/url] [url=https://koji44.com/2021/08/i-will-not-go-home-this-year2/#comment-757]скачать бесплатно на телефон фильм 18 порно[/url] [url=https://www.gaming-updates.com/how-to-cite-an-ebook-in-apa/#comment-5337]порно со сквиртом скачать на телефон[/url] [url=https://www.breakfreebeer.com/2019/03/04/hello-world/#comment-1487]скачать короткое порно на телефон бесплатно[/url] [url=https://theministry.org/hello-world/#comment-52358]секс порно мама скачать телефон[/url] [url=http://runnershorts.com/2018/03/08/the-biggest-reason-you-arent-improving/#comment-41038]скачать порно на телефон две[/url] [url=https://dichvuhuyhoang.vn/phun-thuoc-khu-trung-1/?comment=5418730#comment]скачать на телефон порно самое хорошие[/url] 7ab000c
Lexorra
Все о программе XRumer для Windows https://t.me/progon_sajta_hrumerom/11 Будем рады предложить лучшие условия на покупку легендарного Хрумера
Glenn
paper writing services for college students someone to write my paper pay people to write papers
Rocky
Thanks for finally writing about >Heart Disease Demo: How much risk for a single patient has - Dossiers <Liked it!
Finlay
We are a group of volunteers and starting a new scheme in our community. Your web site provided us with valuable info to work on. You have done an impressive job and our entire community will be thankful to you.
Dominga
Hi there, for all time i used to check website posts here in the early hours in the dawn, for the reason that i enjoy to gain knowledge of more and more.
Dollie
If you wish for to get a good deal from this piece of writing then you have to apply these techniques to your won webpage.
Shirley
Greetings! This is my first visit to your blog!
We are a team of volunteers and starting a new project in a community in the same niche. Your blog provided us useful information to work on. You have done a wonderful job!
Alica
This is a topic which is near to my heart… Cheers!
Where are your contact details though?
Alena
I enjoy what you guys are usually up too. This sort of clever work and coverage! Keep up the awesome works guys I’ve incorporated you guys to my blogroll.
Viola
No matter if some one searches for his vital thing, so he/she wishes to be available that in detail, therefore that thing is maintained over here.
Shenna
Hi there! Do yyou know if tney make anyy plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any tips?
iHerb Code
Where you so for a long time were gone?
Tammara
order custom paper writing services for college papers pay someone to do my paper
Xrumskovs
Для чего можно применять прогоны хрумером? https://t.me/progon_sajta_hrumerom Это один из типов автосабмиттеров, который автономно перебирает ресурсы из базы данных и автоматически размещает на них ссылки, объявления, создает элементарные посты в переписке с пользователями на форумах.
FreeSpins
Не знаете, как получить ежедневные награды - бесплатные вращения: https://t.me/besplatnie_vrashenija Бесплатные вращения или фриспины - самый популярный бонус за регистрацию для новых игроков, который исользуется все чаще для привлечения новой аудитрии.
Boyd
This site was… how do I say it? Relevant!! Finally I’ve found something which helped me.
Many thanks!
Seoshnik
Если обычный текст пишется с учётом того, как продавец хотел бы представить свой товар, то SEO-текст берёт в расчёт алгоритмы, по которым поисковики «считывают» наиболее релевантный контент: https://telegra.ph/CHto-takoe-SEO-test-10-20 Каждый текст рассчитан под запросы целевой аудитории.
Ezra
If you’d like a a lot more detailed guide, check out the ‘How to Get Started’ section earlier in this post.
Antonia
Berikut kurang lebih tips ini. Salah satu penyebab orang2 dapat terjerat di masalah perjudian online dikarenakan daerah sekeliling atau pergaulan dengan cela. Begitu anda bergabung secara daerah sekeliling orang2 dengan gemar tampil judi, oleh karena itu amat besar prospek kalau anda hendak terbawa buat mencoba permainan judi togel online ini. Cobalah buat berkumpul secara orang-orang dengan kiranya mempunyai mindset eksplisit, sehingga siap menghindarkan kamu dibanding godaan judi online serta membentuk watak anda menjadi lebih baik.
Mike
Aw, this was a very nice post. Finding the time and actual effort to make a very good article… but what can I say… I hesitate a whole lot and don’t seem to get anything done.
Pasquale
Hello, I enjoy reading all of your article post. I wanted to write a little comment to support you.
Melanie
Hi there, after reading this amazing paragraph i am also delighted to share my familiarity here with friends.
Karry
Because the admin of this website is working, no uncertainty very shortly it will be well-known, due to its quality contents.
Raul
I want to to thank you for this wonderful read!! I certainly enjoyed every bit of it. I’ve got you bookmarked to look at new things you post…
Leilani
Greetings, I believe your site could possibly be having web browser compatibility issues. Whenever I look at your website in Safari, it looks fine but when opening in IE, it’s got some overlapping issues. I merely wanted to give you a quick heads up! Aside from that, excellent site!
Madelaine
Woah! I’m really digging the template/theme of this blog.
It’s simple, yet effective. A lot of times it’s very difficult to get that “perfect balance” between usability and appearance. I must say you have done a fantastic job with this. Additionally, the blog loads super fast for me on Firefox. Excellent Blog!
Mel
Magnificent web site. Lots of helpful info here.
I am sending it to some pals ans additionally sharing in delicious.
And certainly, thank you to your sweat!
Aleida
Everyone loves what you guys are up too. Such clever work and coverage! Keep up the fantastic works guys I’ve incorporated you guys to blogroll.
Ali
Lilian
People will fill it with speculation that, yes, can be sexist and judgmental and additional, but also simply curious.
Alisia
In April 2020, Florence Pugh, Shia LaBeouf and Chris Pine have been added to the solid of the movie, with Dakota Johnson joining the next month.
Marcy
Perhaps as a result of Wilde wished to film a cool chase scene with old-school cars.
FrankieTrite
Dokąd przeglądać obrazy online pro bonusowo? Ewidencja najszczęśliwszych okolic 2022!
Którykolwiek z nas ubóstwia dążyć do kina azaliż obserwować slajdy online w necie. W teraźniejszych niewielu latach w Polsce wielce rozbudowała się kontroferta tudzież baza obrazów przystępnych wewnątrz poręczeniem netu. Stacjonarną wartość zagrała w bieżącym postęp serwów VOD – progres Netflixa, Player albo CDA Premium. Wszelki spośród bieżących dzienników sprzedaję groźbę poznawanie najnowszych filmów krzew limitu w formie HD na smartfonie, tablecie albo komputerze. Starczy wjazd do netu oraz możemy badać smakuj wchłonąć najdroższy slajd na pecet, który przejrzymy poniewczasie np. w sezonie peregrynacyj nałogiem.
Zwykłe serwisy VOD łatwe w Polsce! Nurt dzienników VOD w Polsce dużo wyrównał. Zbytnio sceną Netflixa, którego kontroferta stanowi najlepsza ze całych dzienników, sprawia się on dodatkowo najtrudniejszą sławą. Nieopodal zbytnio przedtem kładzie się HBO GO, Player albo TVP VOD. Dorosłym upodobaniem syci się ponad konkretne również cenione CDA, z niechwiejnego etapu w prywatnej infrastrukturze CDA Premium ujawniają wewnątrz nikłą taryfą przepisowe półfabrykaty audiowizualne – infrastruktura filmów online bez kresu dzieli nuże nieomal 6300 placówki. Także jest w czym wyłączać i co obserwować. Od oddanego frazeologizmu o osobistą wartość z celuloidami online zabiega płaszczyzna Disney+ spośród slajdami tudzież blagi Walta Disneya, jakiej kolekcja stanowi tak okazała. Facebook tudzież zaciekawił się streamingiem zajść ruchowych na mosso także odtwarzaniem długofalowych negatywów online – w 2017 roku w STANACH zadebiutowała architektura Facebook Watch. W Polsce premierę nosiła w 2018 roku zbieżnie jakże w epizodziku Disney+.
Karty spośród celuloidami online nadto bezsensownie oraz bez zakresu – 2022 Gwoli wielu niewiast kontroferta serwów VOD jest licytacja wadliwa dodatkowo brakuje najefektowniejszych dyplomata negatywów. Zamierzają się na osłuchiwanie filmików online wewnątrz bezskutecznie w okrutniejszej w kondycji na krawędziach, które eksponują nagminnie tajne wykopuje przedmiotów audiowizualnych. Opukiwanie ich nie klanowi uszczuplenie prawdomówna autorskiego aczkolwiek udostępnianie stanowi wprzódy podejrzane, nadmiernie co szantażuje represja kary akceptuj porwanie samodzielności.
https://vod360.pl https://onlinefilmy.pl https://mojseans.pl https://prawdziwefilmy.pl https://kino-man.pl https://nieziemskiefilmy.pl https://filihd.pl https://kiwivod.pl https://zobaczfilmy.pl https://zalukaj-film.pl
PatrickJaida
Use this section to talk about any relevant experience and qualifications. Courses may include. Although the project work and sports activity can give you additional marks, it would be not enough to get the desired course. [url=https://bettyjowrites.com]have a peek at these guys[/url]
CharlesJeono
порно ххх анал большие https://zhopkin.club/ порно фильмы 2021 года [url=https://zhopkin.club/]порно[/url] порна ххх
[img]https://zhopkin.club/picture/Tri-podrugi-v-multiashnykh-kostiumakh-rabotaiut-po-vyzovu-i-ublazhaiut-muzhskie-chleny.jpg[/img]
[url=http://tiwaha.cocolog-nifty.com/blog/2012/04/tokyomx33113-e1.html]ххх порно учительница[/url] [url=https://www.shelbyvillechristianfellowship.com/psalms-a-tool-from-god/#comment-4672]ххх азия порно фильм[/url] [url=https://365dairy.blog.ss-blog.jp/2014-08-01?comment_success=2022-10-22T12:49:30&time=1666410570]порно ххх full[/url] [url=https://www.portablegenerator.co/best-firman-generator/#comment-277016]большая коллекция порно фото[/url] [url=http://hecho.s41.xrea.com/bell/]смотреть фильмы порно хуже[/url] [url=https://kovitechnologies.com/hello-world/#comment-34992]порно душе фильмы ххх[/url] [url=http://hotel-kleopatra.ru/index.php/reviews]смотреть гей порно фильмы[/url] [url=http://blog.blockgold.ee/boris-johnson-impulsa-la-subida-del-oro/#comment-17445]фильмы для взрослых ххх порно скачать[/url] [url=http://climatizacion.ru/guestbook/index.php]порно фильмы 80 годов[/url] [url=http://tokyo-nomunomu.air-nifty.com/blog/2022/03/post-b6f467.html]японские полнометражные порно фильмы[/url] f8_56d2
Senaida
It’s additionally a backdoor pilot for the Justice Society featuring Pierce Brosnan as Doctor Fate, Noah Centineo as Atom Smasher, Quintessa Swindell as Cyclone and Aldis Hodge as Hawkman.
Hildegarde
Shazam learns of this and strips Adam of his powers, encasing them in a mystical scarab necklace.
prosmirwog
Наше предложение выгодно не только тем, что в нем представлены все водительские категории удостоверений. Главное – что такая покупка абсолютно безопасна. Мы оформляем права исключительно на бланках установленного государственного образца.
[url=https://habarovsk.online-prava.com/][img]https://yaroslavl.online-prava.com/baner.jpg[/img][/url] https://nizhnij-novgorod.online-prava.com/spectekhnika/prava-na-traktor.html https://orenburg.online-prava.com/prava/kategoriya-e.html
[url=https://tomsk.online-prava.com/otzyvy.html] купить водительские права с доставкой [/url] [url=https://novosibirsk.online-prava.com/prava/kategoriya-b.html] купить права водительские fort [/url]
купить оригинальный права где купить права цена получение прав купить
Michaeltoobe
How to [url=http://77pro.org/13-fix-damaged-photoshop-psd.html]fix Photoshop[/url] PSD file.
Arnette
Guess the exchange rate, bitcoin and get money. Start with $10 and you can earn up to $1000 in a day, see how Here
Oliver
Временная регистрация в Санкт-Петербурге
Heath
My partner and I absolutely love your blog and find many of your post’s to be exactly I’m looking for. Do you offer guest writers to write content available for you? I wouldn’t mind writing a post or elaborating on some of the subjects you write concerning here. Again, awesome web site!
Garland
I blog quite often and I truly appreciate your content. This article has really peaked my interest. I will bookmark your blog and keep checking for new information about once per week. I opted in for your Feed as well.
Delmar
No matter if some one searches for his necessary thing, therefore he/she wishes to be available that in detail, thus that thing is maintained over here.
Isaac
Thanks for sharing your thoughts on saudi video on us president. Regards
Joy
It’s really very complex in this busy life to listen news on TV, thus I only use internet for that reason, and obtain the newest information.
Dora
I just could not go away your web site prior to suggesting that I extremely enjoyed the standard information a person provide on your visitors? Is going to be again frequently to check up on new posts
Sienna
Yesterday, while I was at work, my cousin stole my apple ipad and tested to see if it can survive a twenty five foot drop, just so she can be a youtube sensation. My iPad is now destroyed and she has 83 views. I know this is completely off topic but I had to share it with someone!
Theron
After looking into a number of the articles on your site, I seriously like your technique of blogging. I added it to my bookmark webpage list and will be checking back soon. Take a look at my web site as well and let me know what you think.
Fredericka
Печи для бани купить в Санкт-Петербурге
Ellie
Good way of telling, and pleasant article to take information concerning my presentation subject, which i am going to convey in institution of higher education.
Margart
Hello it’s me, I am also visiting this web site on a regular basis, this web page is really nice and the viewers are truly sharing pleasant thoughts.
Oliver
Thanks in support of sharing such a nice thinking, piece of writing is fastidious, thats why i have read it fully
Jame
Hello, after reading this amazing article i am as well happy to share my experience here with mates.
Terry
Whoa! This blog looks just like my old one!
It’s on a completely different topic but it has pretty much the same layout and design. Outstanding choice of colors!
Sarah
Does your site have a contact page? I’m having trouble locating it but, I’d like to shoot you an email.
I’ve got some recommendations for your blog you might be interested in hearing. Either way, great site and I look forward to seeing it improve over time.
Ezekiel
Hi! I could have sworn I’ve been to this site before but after browsing through some of the post I realized it’s new to me. Nonetheless, I’m definitely happy I found it and I’ll be bookmarking and checking back often!
Seth
Currently it seems like Wordpress is the best blogging platform out there right now. (from what I’ve read) Is that what you are using on your blog?
Brittny
I do not even know how I ended up here, but I thought this post was good. I do not know who you are but certainly you’re going to a famous blogger if you aren’t already ;) Cheers!
Justinhet
Ищешь где почитать про [url=https://mir74.ru]новости катав[/url]? Много информации об этои есть на сайте Мир74!
Lowell
Excellent blog here! Additionally your web site quite a bit up very fast! What host are you the use of? Can I get your associate link on your host? I wish my website loaded up as quickly as yours lol
Greg
Hi to all, as I am in fact eager of reading this webpage’s post to be updated daily. It carries nice data.
Kiara
great publish, very informative. I’m wondering why the opposite experts of this sector don’t understand this. You should proceed your writing. I am confident, you’ve a huge readers’ base already!
Luigi
Spot on with this write-up, I seriously think this site needs a lot more attention. I’ll probably be back again to see more, thanks for the advice!
Kristian
Greetings! I know this is kinda off topic but I’d figured I’d ask. Would you be interested in trading links or maybe guest authoring a blog article or vice-versa? My site goes over a lot of the same subjects as yours and I believe we could greatly benefit from each other. If you’re interested feel free to shoot me an email. I look forward to hearing from you!
Awesome blog by the way!
Georgina
Pretty section of content. I just stumbled upon your weblog and in accession capital to assert that I get actually enjoyed account your blog posts. Any way I’ll be subscribing to your augment and even I achievement you access consistently fast.
Tandy
I like what you guys tend to be up too. This sort of clever work and exposure! Keep up the superb works guys I’ve added you guys to my own blogroll.
Virtual Local Numbers
Thanks for an explanation. I did not know it.
idtqmqcp
erect plus pills
[url=https://viagaratime.com/]order generic viagra[/url]
safe erectile dysfunction pills
what is the chemical name for viagra [url=https://viagaratime.com/]online generic viagra[/url] can i use viagra daily
Willie
Relax and enjoy one of the Professors signature cocktails as you await your journey through time.
Lasonya
Then a non-linear version of escape rooms has more complexity than the linear types.
RonaldDus
The percentage remaining declines in an approximately exponential manner, similar to the decay curve for a radioactive chemical element the time for the number to decline by 50 is roughly the same, regardless of the initial num. Examine the tops and undersides of leaves for pests or discoloration spider mites live on the underside of leaves as well as stalks and branches. And we couldn t be happier with the resulting Indica hybrid a top-notch strain with many of the original traits that triumphs everywhere and that everyone identifies as an old-school classic. [url=https://freesound.org/people/maadmacs/sounds/388391/]https://freesound.org/people/maadmacs/sounds/388391/[/url]
Homer
Very nice article, totally what I was looking for.
SeoRuns
Все лучшие базы для Хрумака на сайте https://www.pinterest.de/seoruns/ . Оцените мой Pintetest сайт, спасибо за внимание :)
Scot
Incredible! This blog looks just like my old one! It’s on a entirely different topic but it has pretty much the same layout and design. Superb choice of colors!
Archie
Hey there! Quick question that’s completely off topic. Do you know how to make your site mobile friendly? My weblog looks weird when viewing from my iphone 4. I’m trying to find a theme or plugin that might be able to correct this problem. If you have any suggestions, please share. Thank you!
Wendy
Hey I know this is off topic but I was wondering if you knew of any widgets I could add to my blog that automatically tweet my newest twitter updates. I’ve been looking for a plug-in like this for quite some time and was hoping maybe you would have some experience with something like this. Please let me know if you run into anything. I truly enjoy reading your blog and I look forward to your new updates.
Rob
Greetings from Ohio! I’m bored to tears at work so I decided to browse your blog on my iphone during lunch break. I love the knowledge you provide here and can’t wait to take a look when I get home. I’m shocked at how fast your blog loaded on my mobile .. I’m not even using WIFI, just 3G .. Anyhow, amazing site!
Hollie
Hi, I do think this is an excellent website. I stumbledupon it ;) I am going to return yet again since i have book marked it. Money and freedom is the best way to change, may you be rich and continue to help other people.
Bridgett
If you are going for most excellent contents like myself, only go to see this site every day as it gives quality contents, thanks
Corey
Hello! I’ve been following your web site for a long time now and finally got the courage to go ahead and give you a shout out from Kingwood Tx! Just wanted to mention keep up the excellent job!
Willard
Hi! I’ve been reading your weblog for some time now and finally got the bravery to go ahead and give you a shout out from Porter Texas! Just wanted to mention keep up the fantastic job!
Fredericka
whoah this blog is fantastic i love reading your articles. Keep up the good work! You recognize, lots of people are looking round for this information, you can aid them greatly.
Wyatt
Corporations, restricted liability corporations, and limited partnerships in Georgia are fashioned by submitting with the Corporations Division.
datafastproxiespx01
IPv6 Proxy to Resolve, Highest Quality IPv6 Proxy! DataFast Proxies IPv6 Proxy, Definitive Solution in IPv6 Proxy Agent (100% Anonymous, Undetectable at L2 and L4 OSI Model layers, no DNS leak, no header leak)!
Highest Quality IPv6 Proxy so you get the highest results Solving Google reCAPTCHA v1, v2 is v3 using XEvil or Capmonster!
DataFast Proxies | Definitive Solution in IPv6 Proxy! https://datafastproxies.com/
Contact: https://datafastproxies.com/contact/
Aiden
You ought to take part in a contest for one of the highest quality blogs on the web. I most certainly will recommend this web site!
Sommer
It’s very effortless to find out any topic on net as compared to textbooks, as I found this article at this web site.
Odell
Everything is very open with a very clear description of the issues.
It was truly informative. Your website is useful. Thanks for sharing!
Latisha
For hottest news you have to visit world-wide-web and on the web I found this web page as a best web site for most recent updates.
Rebecca
You could certainly see your skills in the work you write. The sector hopes for more passionate writers such as you who aren’t afraid to say how they believe. At all times follow your heart.
Alfonzo
Howdy! I know this is kinda off topic but I was wondering which blog platform are you using for this site? I’m getting fed up of Wordpress because I’ve had issues with hackers and I’m looking at options for another platform. I would be awesome if you could point me in the direction of a good platform.
August
Incredible story there. What happened after? Thanks!
Eugene
Hello to all, how is the whole thing, I think every one is getting more from this web page, and your views are pleasant in favor of new viewers.
Clayton
Wow, that’s what I was looking for, what a data! present here at this webpage, thanks admin of this web page.
Gretta
I do accept as true with all the ideas you have offered for your post. They are very convincing and can certainly work. Nonetheless, the posts are very brief for newbies. Could you please extend them a little from next time? Thank you for the post.
Paula
Touche. Sound arguments. Keep up the amazing effort.
Milton
Why viewers still make use of to read news papers when in this technological globe the whole thing is available on net?
Hayley
It’s an awesome paragraph in support of all the internet visitors; they will get benefit from it I am sure.
Andrew
Your mode of describing the whole thing in this article is actually fastidious, every one can easily know it, Thanks a lot.
Enriqueta
Its like you read my mind! You seem to know so much about this, like you wrote the book in it or something. I think that you can do with some pics to drive the message home a bit, but other than that, this is excellent blog. An excellent read. I will definitely be back.
Lucy
It’s awesome in support of me to have a site, which is beneficial in favor of my know-how. thanks admin
DwainLauts
Bravo, what necessary phrase…, a magnificent idea https://hotmilfmoms.info https://imagine-dream.com
Chad
I must thank you for the efforts you have put in writing this website. I am hoping to check out the same high-grade blog posts by you in the future as well. In fact, your creative writing abilities has encouraged me to get my very own website now ;)
Lillian
Great article. I will be experiencing some of these issues as well..
Jude
Please let me know if you’re looking for a writer for your site. You have some really great articles and I feel I would be a good asset. If you ever want to take some of the load off, I’d love to write some material for your blog in exchange for a link back to mine. Please blast me an e-mail if interested. Many thanks!
Wyatt
I’ve been browsing on-line more than 3 hours lately, but I never found any fascinating article like yours. It is pretty worth sufficient for me. In my opinion, if all site owners and bloggers made good content material as you did, the net might be much more useful than ever before.
Foster
My family members every time say that I am killing my time here at web, but I know I am getting know-how all the time by reading such fastidious posts.
Bonny
I truly love your blog.. Excellent colors & theme. Did you develop this site yourself? Please reply back as I’m trying to create my own site and want to know where you got this from or exactly what the theme is called. Kudos!
Jacqueline
Thank you a lot for sharing this with all people you actually know what you’re speaking about! Bookmarked. Kindly also seek advice from my site =). We may have a hyperlink exchange contract between us
Janessa
Good replies in return of this question with genuine arguments and describing the whole thing concerning that.
Venus
In his free time he loves going for runs and enjoying together with his beagle Butterscotch.
discount online pharmacy
online pharmacy no peescription https://greatcanadianpharmacies.com/ [url=https://greatcanadianpharmacies.com/]legitimate online canadian pharmacies[/url]
Maggie
Ιf some one desires expert vіew about blogging thеn i recommend him/hеr tⲟ go to see this website, Keep uр thе fastidious ᴡork.
Aleida
Awesome! Its actually awesome article, I have got much clear idea regarding from this post.
Tracy
I was suggested this website by means of my cousin. I am now not certain whether or not this publish is written by way of him as no one else recognize such targeted about my problem.
You are incredible! Thanks!
Verlene
Hello! I know this is kind of off topic but I was wondering if you knew where I could find a captcha plugin for my comment form? I’m using the same blog platform as yours and I’m having trouble finding one? Thanks a lot!
Lelia
I wanted to thank you for this very good read!!
I certainly enjoyed every bit of it. I’ve got you book marked to check out new stuff you post…
Ute
Hello there, You have done a great job. I’ll certainly digg it and personally recommend to my friends. I’m confident they’ll be benefited from this website.
Lori
When someone writes an post he/she keeps the image of a user in his/her mind that how a user can know it. Thus that’s why this post is amazing. Thanks!
Sammy
I needed to thank you for this excellent read!! I definitely loved every little bit of it. I have got you book-marked to check out new things you post…
Fredric
Excellent post. I am going through a few of these issues as well..
Stormy
I read this piece of writing completely concerning the resemblance of hottest and earlier technologies, it’s amazing article.
Corey
I will immediately grasp your rss feed as I can’t in finding your email subscription link or e-newsletter service. Do you have any? Kindly allow me understand so that I may subscribe. Thanks.
Isiah
I’m gone to inform my little brother, that he should also visit this website on regular basis to obtain updated from most up-to-date information.
Pasquale
Thankfulness to my father who stated to me concerning this web site, this webpage is in fact awesome.
Ronaldgam
Most professional courting services expect you to pay out a payment to sign up, but you will find free of charge ladies in the area using a straightforward internet search. Most free of charge web sites will assist you to talk to these girls for free. Even so, if you are looking to meet these ladies face-to-face, you will need to shell out a charge. While many skip the games florence free web sites provide the opportunity to talk to these ladies, you must not expect any kind of those to divulge your current email address or other private information for you.
Maybelle
Does your blog have a contact page? I’m having a tough time locating it but, I’d like to shoot you an e-mail.
I’ve got some recommendations for your blog you might be interested in hearing. Either way, great site and I look forward to seeing it improve over time.
Sammy
Промокод 1xbet
pesy
Таможенный агент «ВЭД ЛАЙН» выступает в качестве ключевого поставщика в международной деятельности с Китаем. Наша команда хранит богатый опыт в этой действенности, так как мы работаем на этом рынке много лет. «ВЭД ЛАЙН» имеет квалифицированных менеджеров по работе с импортными поставками, в том числе опытных менеджеров по сотрудничеству с покупателями. http://www.scampatrol.org/tools/whois.php?domain=ved-line.ru
[url=https://www.carillonmons.be/en/guestbook/]Логистика и таможенное оформление[/url] 4b7ab00
Felipa
I think the admin of this website is in fact working hard in support of his web site, for the reason that here every information is quality based material.
Fern
Saved as a favorite, I like your site!
Flossie
You can definitely see your enthusiasm within the work you write. The arena hopes for even more passionate writers like you who are not afraid to mention how they believe. At all times follow your heart.
Debora
I’ve been surfing online more than 3 hours today, yet I never found any interesting article like yours. It’s pretty worth enough for me. In my view, if all website owners and bloggers made good content as you did, the web will be much more useful than ever before.
Davidtek
Hello! [url=http://stromectolrf.top/]ivermectin oral 0 8[/url] beneficial internet site http://stromectolrf.online
Aiden
For hottest news you have to pay a quick visit world wide web and on the web I found this web site as a best site for newest updates.
Francisco
I pay a visit every day some blogs and blogs to read content, but this weblog provides quality based writing.
Ethel
I must thank you for the efforts you have put in penning this site. I’m hoping to view the same high-grade content from you later on as well. In fact, your creative writing abilities has inspired me to get my own, personal blog now ;)
Elliot
інструмент
Lamont
Directed by Shi and produced by Lindsey Collins, Disney and Pixar’s “Turning Red” is now streaming exclusively on Disney+.
Inge
Hello everyone, it’s my first pay a visit at this web site, and paragraph is truly fruitful for me, keep up posting these types of posts.
Russellcycle
Howdy! [url=http://iscialis.top/]buy cialis 2.5 mg online[/url] very good web page http://iscialis.top
Russellcycle
Hi there! [url=http://iscialis.online/]order generic cialis by phone[/url] very good website http://iscialis.top
Marcia
The name of the town dates from the twelfth century, when a dam was constructed within the Amstel river.
Eugenio
It looks like a parody of a David O. Russell movie - all imitation, no authenticity.
Adelaide
Thanks for any other wonderful post. Where else may anybody get that type of information in such an ideal method of writing?
I’ve a presentation next week, and I am on the look for such information.
Etta
Piece of writing writing іs also a fun, іf yօu кnow afterward үou ⅽan write or else it iѕ difficult to write.
Delia
Thhe resort also has a 1,108 space casino, spa, 11 restaurants, a golf club, andd a evening lounge.
Eusebia
By creating an account, you agree to the Privacy Policyand the Terms and Policies, and to receive email from Rotten Tomatoes and Fandango.
Russellcycle
Hello! [url=http://iscialis.top/]buy cialis ebay[/url] excellent website http://iscialis.top
Blondell
Hi, i read your blog occasionally and i own a similar one and i was just wondering if you get a lot of spam remarks? If so how do you stop it, any plugin or anything you can recommend? I get so much lately it’s driving me insane so any assistance is very much appreciated.
Russellcycle
Hello there! [url=http://iscialis.top/]buy cialis online canada pharmacy[/url] very good web page http://iscialis.top
Tylerkic
Hello! [url=http://pharmacyonlinexp.quest/]canadian prescriptions[/url] beneficial web site http://onlinesxpharmacy.quest
Lynn
Wonderful blog! I found it while searching on Yahoo News. Do you have any suggestions on how to get listed in Yahoo News? I’ve been trying for a while but I never seem to get there! Thanks
Ward
Hmm is anyone else having problems with the images on this blog loading? I’m trying to determine if its a problem on my end or if it’s the blog. Any feedback would be greatly appreciated.
Scot
If you are going for best contents like myself, just go to see this web page daily since it provides feature contents, thanks
Lukas
I think this is one of the most important information for me. And i am glad reading your article. But should remark on some general things, The site style is great, the articles is really nice : D. Good job, cheers
Emmett
Simply desire to say your article is as astounding. The clarity in your post is simply excellent and i can assume you are an expert on this subject. Fine with your permission let me to grab your feed to keep updated with forthcoming post. Thanks a million and please keep up the enjoyable work.
Maxwell
As the admin of this web page is working, no doubt very soon it will be well-known, due to its quality contents.
Trevor
I do consider all of the concepts you’ve offered to your post.
They are very convincing and can definitely work. Nonetheless, the posts are too quick for novices.
May you please extend them a bit from subsequent time? Thank you for the post.
Nila
Hi Dear, are you really visiting this web page on a regular basis, if so then you will absolutely get fastidious knowledge.
Tylerkic
Hello! [url=http://onlinesxpharmacy.quest/]prescription without a doctor’s prescription[/url] very good internet site http://pharmacyonlinexp.quest
Tylerkic
Howdy! [url=http://pharmacyonlinexp.quest/]no prescription pharmacies[/url] good web site http://pharmacyonlinexp.quest
Marvin
When 2020’s movies began to be reshuffled, Uncharted was the first 2021 movie to be moved.
Penny
As for Goose, the cat, we won’t say more about his position to not “express”.
Tylerkic
Hi! [url=http://onlinesxpharmacy.quest/]canadian pharmacies[/url] beneficial site http://onlinesxpharmacy.quest
George
And once once more, we discover ourselves with a bounty of nice work before we’ve even hit the autumn season.
Tylerkic
Hi! [url=http://onlinesxpharmacy.quest/]canada pharmacies prescription drugs[/url] excellent internet site http://pharmacyonlinexp.quest
Tylerkic
Hi there! [url=http://pharmacyonlinexp.quest/]drug prices comparison[/url] very good web site http://onlinesxpharmacy.quest
Tangela
Hi there, this weekend is nice in support of me, because this occasion i am reading this impressive informative post here at my house.
Lora
Hello! I simply wish to offer you a huge thumbs up for the excellent info you have here on this post. I’ll be coming back to your blog for more soon.
Gino
I loved as much as you will receive carried out right here. The sketch is tasteful, your authored subject matter stylish. nonetheless, you command get bought an nervousness over that you wish be delivering the following. unwell unquestionably come more formerly again as exactly the same nearly a lot often inside case you shield this hike.
Hortense
What’s Going down i am new to this, I stumbled upon this I’ve found It absolutely useful and it has helped me out loads. I’m hoping to give a contribution & assist other customers like its helped me. Great job.
Tylerkic
Hi! [url=http://onlinesxpharmacy.quest/]canadian pharmacy[/url] good web page http://pharmacyonlinexp.quest
Tylerkic
Hello! [url=http://onlinesxpharmacy.quest/]drugstore online[/url] excellent web site http://pharmacyonlinexp.quest
Tylerkic
Hi! [url=http://pharmacyonlinexp.quest/]canadapharmacy com[/url] very good site http://pharmacyonlinexp.quest
Joshua
The hilltop headquarters of the Victory Corporation is the long-lasting “volcano house” within the Mojave Desert group of Newberry Springs, California.
Charla
During the process, she sees visions of herself in one other life, as a present-day surgeon named Alice Warren, who lives with the unemployed Jack and struggles to make ends meet.
FrankExign
[url=https://www.casinozru.com/]В индустрии онлайн-гемблинга каждую неделю появляются новые интернет- казино. Многие из них очень похожи друг на друга, а некоторые проекты удивляют необычным дизайном, оригинальными названиями, нестандартными акциями и другими нетипичными особенностями. Разобраться в многообразии новинок поможет Casinoz. Наши обозреватели постоянно мониторят рынок и отслеживают последние новинки. В подробных статьях рассматриваются бонусы, слоты, турниры, джекпоты, игры с живыми дилерами, программы лояльности и VIP-услуги. Также учитывается наличие лицензий, сертификатов и печатей одобрения тематических СМИ. Новые казино не дают скучать любителям азартных игр в интернете, но их огромное количество заставляет пользователей ломать голову над проблемой выбора. Мы подберем для вас честное, надежное и качественное заведение с лучшим перечнем услуг.[/url]
[url=https://www.casinozru.com/]Посещаете ли вы казино ради удовольствия или же считаете игру своей профессией, вам необходим внушительный багаж знаний, без которых придется туго. Чтобы игровой процесс был максимально приятным и эффективным, необходимо разобраться в многих вопросах. На помощь любителям азарта приходит Casinoz. Это бездонный источник важной информации обо всех аспектах игорного бизнеса. Ежедневно его пополняют новые материалы разных жанров: статьи, обзоры, новости, видео, инфографика и так далее. Все они призваны дать новые знания о казино и сопутствующих темах. Не стоит отправляться в плавание по бурным водам гемблинга без надежного партнера. Лучшим компаньоном станет коллективный опыт наших экспертов, авторов, обозревателей и читателей.[/url]
Mitchell
That’s all led to chatter a few possible break up, which might give us a becoming, symmetrical end to the movie’s mythology.
Caryn
It options an ensemble forged that includes Florence Pugh, Harry Styles, Wilde, Gemma Chan, KiKi Layne, Nick Kroll, and Chris Pine.
Selena
To her husband’s displeasure, Alice begins to have doubts about the system of dinners and ballet classes and decides to try to break free.
Deloris
Howdy! This article could not be written any better! Reading through this post reminds me of my previous roommate! He continually kept preaching about this. I am going to send this information to him. Fairly certain he will have a great read. Thank you for sharing!
Jarred
Have you ever thought about creating an e-book or guest authoring on other websites? I have a blog based upon on the same information you discuss and would really like to have you share some stories/information. I know my audience would enjoy your work. If you are even remotely interested, feel free to send me an e-mail.
Brad
Thanks for a marvelous posting! I truly enjoyed reading it, you might be a great author.I will make sure to bookmark your blog and will eventually come back sometime soon. I want to encourage you continue your great writing, have a nice holiday weekend!
Elyse
If some one desires expert view concerning blogging then i advise him/her to visit this webpage, Keep up the pleasant job.
Jon
Appreciating the time and energy you put into your site and detailed information you offer. It’s great to come across a blog every once in a while that isn’t the same out of date rehashed material. Fantastic read!
I’ve bookmarked your site and I’m adding your RSS feeds to my Google account.
Bailey
I really like your blog.. very nice colors & theme.
Did you make this website yourself or did you hire someone to do it for you? Plz respond as I’m looking to construct my own blog and would like to know where u got this from. many thanks
Britt
Hello, I enjoy reading through your article. I wanted to write a little comment to support you.
Michaela
What’s up to all, how is the whole thing, I think every one is getting more from this web page, and your views are pleasant for new viewers.
Clinton
Good post. I learn something totally new and challenging on blogs I stumbleupon everyday. It’s always helpful to read content from other writers and practice a little something from other web sites.
FrankExign
[url=https://affgambler.ru/]продам гашиш[/url]
John
We too have convinced ourselves that certainly ‘the larger the bonus, the much better the betting site’.
FrankExign
[url=https://latestcasinobonuses.me/]Получить дополнительное преимущество в игре и увеличить шансы на крупный занос помогут новые и самые свежие бонусы казино, так как они прибавляют монет к общей сумме банкролла. Наведываясь в данный раздел время от времени или подписавшись на обновления, вы будете всегда находиться в курсе событий на счет актуальных и проходящих в текущий момент бонусных предложений. Бездепы, бонус на депозит, кэшбэк – все это дополнительные средства, продлевающие игровую сессию и вместе с этим прибавляющие возможности для солидных выигрышей. Стоит обратить внимание на вагер. Некоторые бонусы казино предоставляются без отыгрыша или с довольно привлекательными условиями. Также в перечне новых и самых свежих бонусов казино указываются эксклюзивные предложения, которыми могут воспользоваться на специальных условиях только посетители и зарегистрированные участники сообщества ЛатестКазиноБонус.[/url]
Sandypsype
iHerb promo code 5-80 % off » https://code-herb.com/
Alejandra
In 2006, advertisers spent $280 million on social networks.
Social context graph mannequin (SCGM) (Fotakis et al., 2011) contemplating adjacent context of advert is upon the assumption of separable CTRs, and GSP with SCGM has the same problem. Here’s one other scenario for you: You give your boyfriend your Facebook password because he needs that can assist you upload some vacation images.
It’s also possible to e-mail the photographs in your album to anyone with a pc and an e-mail account.
Phishing is a scam through which you receive a faux e-mail that seems to come back from your financial institution, a merchant or an auction Web site. The site aims to help users “set up, share and discover” throughout the yarn artisan group. For example, guidelines could direct users to use a certain tone or language on the location, or they might forbid certain conduct (like harassment or spamming). Facebook publishes any Flixster exercise to the user’s feed, which attracts other customers to join in. The costs rise consecutively for the three different items, which have Intel i9-11900H processors. There are four configurations of the Asus ROG Zephyrus S17 on the Asus webpage, with prices beginning at $2,199.Ninety nine for fashions with a i7-11800H processor.
For the latter, Asus has opted to not place them off the lower periphery of the keyboard.
Edison
It’s actually a cool and helpful piece of information. I’m glad that you just shared this helpful info with us. Please keep us up to date like this. Thank you for sharing.
Deborah
Can you tell us more about this? I’d love to find out some additional information.
Jerry
There’s certainly a great deal to know about this issue.
I love all the points you’ve made.
Ruben
Link exchange is nothing else but it is only placing the other person’s blog link on your page at proper place and other person will also do similar in support of you.
FrankExign
[url=https://stamps-maker.com/]The Best online Stamp constructor Custom Stamps Maker Create Custom Stamps featuring your personalized text and logos quickly and easily at Stamps-Maker.[/url] [url=https://stamps-maker.com/]Feel like a designer and create a rubber stamp template from scratch. You will have access to many functions for editing text, frames, images, etc. Everything is limited only by your imagination.
Don’t want to create a template from scratch? No problem! Just choose the stamp template you like. Change the template you like! Save the layout and download it for your own use..[/url] [url=https://stamps-maker.com/]What result? We are not stamp manufacturers!
We do not sell stamp body, ink or other accessories.
You can download the template in different formats.
You can use a digital stamp for documents or make a rubber stamp from a template.
If you want to get a rubber stamp, then just send the saved file to the nearest stamp manufacturer.[/url] [url=https://stamps-maker.com/]If you need business cards or flyers, then you can use the online designer from PRINTUT company. Over 6000 templates from professional designers. Go to PRINTUT.COM and create your own designs![/url]
Randall
Undeniably believe that which you stated. Your favorite justification appeared to be on the web the easiest thing to be aware of. I say to you, I definitely get annoyed while people think about worries that they just do not know about. You managed to hit the nail upon the top as well as defined out the whole thing without having side-effects , people could take a signal. Will probably be back to get more. Thanks
Harry
I simply couldn’t leave your web site prior to suggesting that I really loved the usual information an individual supply for your visitors? Is gonna be back continuously to inspect new posts
King
I every time emailed this webpage post page to all my contacts, since if like to read it after that my contacts will too.
Octavia
Hello, everything is going well here and ofcourse every one is sharing facts, that’s truly good, keep up writing.
Annett
Somebody necessarily help to make critically articles I might state. This is the very first time I frequented your website page and up to now? I amazed with the research you made to make this actual submit amazing. Excellent task!
Corine
I was extremely pleased to uncover this page. I need to to thank you for ones time just for this fantastic read!! I definitely enjoyed every little bit of it and I have you book marked to look at new information in your website.
Tonja
Hi there! I realize this is somewhat off-topic however I had to ask. Does managing a well-established blog like yours require a large amount of work? I’m completely new to writing a blog however I do write in my journal daily. I’d like to start a blog so I can easily share my experience and feelings online.
Please let me know if you have any kind of recommendations or tips for new aspiring blog owners.
Thankyou!
Bettina
Can I simply say what a relief to find an individual who really knows what they are talking about online. You definitely understand how to bring an issue to light and make it important. More people ought to look at this and understand this side of the story. I was surprised that you’re not more popular since you surely possess the gift.
Georgewrene
Howdy! [url=http://xlppharm.com/]canadian pharmacy domperidone[/url] good internet site http://xlppharm.com
RustyCrymn
Leanna
They expect to offer full entry to Google Play soon. If you are bent on getting a full-fledged pill experience, with entry to every raved-about app and all the bells and whistles, a Nextbook probably is not the only option for you. Pure sine wave inverters produce AC power with the least amount of harmonic distortion and could also be the best choice; but they’re also typically the most expensive. Not to fret. This article lists what we consider to be the 5 best options for listening to CDs in your car for those who only have a cassette player. So listed here are, in no explicit order, the highest 5 choices for playing a CD in your automobile if you solely have a cassette-tape participant. Whether you’re speaking about Instagram, Twitter or Snapchat, social media is a trend that’s right here to remain. Davila, Damina. “Baby Boomers Get Connected with Social Media.” idaconcpts.
Some kits come complete with a mounting bracket that means that you can fasten your portable CD participant securely inside your car.
Perhaps the simplest methodology for listening to a CD player in a car without an in-sprint CD participant is by means of an FM modulator.
Georgewrene
Hello! [url=http://xlppharm.com/]free pharmacy tech training online[/url] very good web site http://xlppharm.com
FrankExign
[url=https://stamps-maker.com/]The Best online Stamp constructor Custom Stamps Maker Create Custom Stamps featuring your personalized text and logos quickly and easily at Stamps-Maker.[/url] [url=https://stamps-maker.com/]Feel like a designer and create a rubber stamp template from scratch. You will have access to many functions for editing text, frames, images, etc. Everything is limited only by your imagination.
Don’t want to create a template from scratch? No problem! Just choose the stamp template you like. Change the template you like! Save the layout and download it for your own use..[/url] [url=https://stamps-maker.com/]What result? We are not stamp manufacturers!
We do not sell stamp body, ink or other accessories.
You can download the template in different formats.
You can use a digital stamp for documents or make a rubber stamp from a template.
If you want to get a rubber stamp, then just send the saved file to the nearest stamp manufacturer.[/url] [url=https://stamps-maker.com/]If you need business cards or flyers, then you can use the online designer from PRINTUT company. Over 6000 templates from professional designers. Go to PRINTUT.COM and create your own designs![/url]
Christen
Hi! Do you know if they make any plugins to protect against hackers?
I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
Georgewrene
Hello there! [url=http://xlppharm.com/]walmart pharmacy online refills[/url] great website http://xlppharm.com
FrankExign
[url=https://www.casinozru.com/]Как выбрать лучшее интернет-казино? Задача не из простых, учитывая огромное количество игорных порталов в сети. Можно самостоятельно проверять лицензии, читать обзоры, изучать отзывы реальных клиентов, сравнивать игровые автоматы, углубляться в особенности бонусной программы. Но гораздо проще довериться профессионалам, составляющим рейтинг онлайн- казино. На Casinoz список наиболее качественных игорных заведений формируется на основе объективных характеристик, мнений пользователей, экспертных оценок и дополнительных факторов. Комплексный подход дает возможность отобрать действительно лучшие казино, достойные внимания самых требовательных пользователей. Вы можете также оставить свой отзыв, который будет учтен при обновлении рейтинга.[/url]
[url=https://www.casinozru.com/]Casinoz не оставляет без внимания ни один новый слот, выпущенный известным разработчиком софта. На портале опубликованы тысячи обзоров игровых автоматов большинства популярных производителей. К ним прилагаются видео с демонстрацией возможностей и демо-версия, позволяющая испытать видеослот бесплатно. Лучшие азартные игры NetEnt, Microgaming, Betsoft, Yggdrasil и других знаменитых брендов обладают потрясающим дизайном, невероятной анимацией, уникальными бонусами и оригинальными возможностями геймплея. Многие новинки – это настоящие шедевры, которые задают тренды и становятся объектами для подражания. Обзоры новых игровых аппаратов познакомят вас с особенностями интерфейса, нюансами правил и ключевыми характеристиками моделей. Вы сможете протестировать их, посмотреть видео-обзор и решить, стоит ли запускать слот на деньги.[/url]
Georgewrene
Hello! [url=http://xlppharm.com/]walmart pharmacy refill online[/url] good internet site http://xlppharm.com
FrankExign
[url=https://www.casinozru.com/]В жизни Casinoz активно участвуют многие неравнодушные пользователи. Они не только читают публикации, но и ведут блоги о казино в специальном разделе портала. В числе постоянных авторов много интересных людей: сотрудников казино, профессиональных игроков, любителей покера, экспертов букмекерских ставок и специалистов в других направлениях. Их дневники – это кладезь народной мудрости, которой они щедро делятся со всеми единомышленниками. Вам нужны практические советы, объективные мнения и четкие рекомендации? Тогда следите за обновлениями в блогах об азартных играх. Вы также можете завести тематический дневник, чтобы поведать миру о своих взглядах на разные вопросы игорного бизнеса.[/url] [url=https://www.casinozru.com/]Если вы азартны и не мыслите жизни без общения, становитесь участником форума Casinoz. Он открыт для всех желающих и охватывают все самые популярные темы и жанры. На форуме вы встретите игроков, сотрудников офлайновых казино, операторов гемблинга, обозревателей портала, внештатных авторов и многих других интересных людей. Вы сможете напрямую поговорить с официальными представителями интернет- казино, задать вопрос профессиональным игрокам, узнать мнение общественности о новых казино и получить другую информацию.[/url]
[url=https://www.casinozru.com/]Как выбрать лучшее интернет-казино? Задача не из простых, учитывая огромное количество игорных порталов в сети. Можно самостоятельно проверять лицензии, читать обзоры, изучать отзывы реальных клиентов, сравнивать игровые автоматы, углубляться в особенности бонусной программы. Но гораздо проще довериться профессионалам, составляющим рейтинг онлайн- казино. На Casinoz список наиболее качественных игорных заведений формируется на основе объективных характеристик, мнений пользователей, экспертных оценок и дополнительных факторов. Комплексный подход дает возможность отобрать действительно лучшие казино, достойные внимания самых требовательных пользователей. Вы можете также оставить свой отзыв, который будет учтен при обновлении рейтинга.[/url]
Harrison
It’s no wonder that he shaped the Invaders throughout World War II to defend New York from the Nazis.
Vito
The movie was included in a sizzle reel shown ahead of screenings throughout National Cinema Day that highlighted upcoming movies from various studios.
Ramon
The sequel will also introduce fans to Huerta as Namor, Thorne as Ironheart, Michaela Coel as Aneka, Mabel Cadena as Namora, María Mercedes Coroy as Princess Fen, Alex Livinalli as Attuma, and extra.
Mari
Feige and other executives at Marvel Studios have been additionally unaware of Boseman’s illness.
Corey
He was expected to start writing the sequel in 2019, forward of a planned filming begin in late 2019 or early 2020.
FrankExign
[url=https://affgambler.ru/]продам героин[/url]
Ken
Production resumed by mid-January 2022 and wrapped in late March in Puerto Rico.
MichaelVes
Hi there! [url=http://xuonlinepharmacy.com/]canadian pharmacies selling cialis[/url] very good internet site http://xuonlinepharmacy.com
FrankExign
[url=https://affgambler.ru/]продам кокоин[/url]
Juli
Hi, I read your blogs daily. Your writing style is awesome, keep doing what you’re doing!
MichaelVes
Hello there! [url=http://xuonlinepharmacy.com/]online pharmacy 365 pills[/url] beneficial web site http://xuonlinepharmacy.com
GedInsof
amateur video porno
http://www.ptech.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://sfga.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories
Shad
From Oscar-nominated visionary filmmaker Baz Luhrmann comes Warner Bros.
Antony
Despite this, forty seven states are attempting to move laws making it more difficult to vote, claiming that they’re doing it to keep away from the notion of fraud. They’re doing it as a result of there’s cash in it. Finally, what may be most irritating about all of that is that the electronic platforms hosting this fraud, from Facebook and Twitter to eBay, seem to be doing virtually nothing about it.
While the No. 2 member of the Disinformation Dozen may be the perennially sorry excuse and long established youngster-killer Robert Kennedy Jr., the third slot belongs to Ty & Charlene Bollinger. The Disinformation Dozen could dominate false claims circulating on social media, but they are removed from alone. So are most of the others in the Disinformation Dozen. At the moment, automobile sharing exists in lots of of cities across more than a dozen international locations, with more doubtless to come back. The automobile isn’t roadworthy but since a hybrid engine (made of metal) still has be designed, not to say security exams should be carried out. A research carried out by the University of Virginia found that of the highest one hundred fifty functions on Facebook, ninety p.c have been given entry to info they did not want in order for the app to operate.
MichaelVes
Hello! [url=http://xuonlinepharmacy.com/]prescription drugs discount[/url] good web site http://xuonlinepharmacy.com
FrankExign
[url=https://latestcasinobonuses.me/]Онлайн казино никогда не покажут себя с негативной стороны. Даже, если будет предоставлен прямой факт, говорящий о нечестных действиях со стороны игорного заведения, все равно, представитель покроет все туманом или сошлется на неправомерные действия со стороны клиента. При этом многие (если не подавляющее большинство(!)) обзоры и отзывы об игорных площадках в интернете проплачены и практически не отражают действительного качества обслуживания заведения. Здесь на помощь придут реальные отзывы игроков о казино, опубликованные на сайте Латеста. В них посетители азартных ресурсов рассказывают о полученных впечатлениях, об удобстве пользования игорным сайтом, насколько широк ассортимент представленных игр и, конечно, как обстояли дела с выводом средств: были ли попытки не выплатить или необоснованные задержки с перечислением, чтобы игрок начал новую сессию, но уже не такую успешную и все проиграл. Среди реальных отзывов игроков о казино можно найти информацию об отыгрыше бездепозитных бонусов в казино, как затем происходил процесс вывода, требовало ли заведение верификацию, внесение первого депозита и за какой период прошло перечисление. Оставляя свои отзывы, лудоман помогает собрату лудоману не тратить свое время и деньги на игру в нечестном казино.[/url]
Alton
Подробные условия этих и других акций ищем в соответствующем разделе на сайте букмекера Mostbet.
GedInsof
hot milf sexy
http://tdmes.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories http://www.icbpro.com/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
MichaelVes
Hi there! [url=http://xuonlinepharmacy.com/]precription drugs from canada[/url] beneficial web site http://xuonlinepharmacy.com
GedInsof
raped milf
http://xn–b1aktdfh3fwa.xn–p1ai/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://www.infocreative.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories
StevenBexia
Hello there! [url=http://stromectolxlp.com/]stromectol online[/url] excellent web page http://stromectolxlp.com
Clay
Пластиковые окна купить в Москве
GedInsof
porno very teens
http://www.aelita.biz/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories http://ug-gsm.com/bitrix/redirect.php?event1=&event2=&event3=&goto=https://tubesweet.xyz/categories
Henrietta
I read this piece of writing fully concerning the comparison of hottest and earlier technologies, it’s remarkable article.
StevenBexia
Hello there! [url=http://stromectolxlp.com/]buy stromectol australia[/url] beneficial website http://stromectolxlp.com
Angelica
Hello! I know this is kind of off topic but I was wondering which blog platform are you using for this site? I’m getting tired of Wordpress because I’ve had issues with hackers and I’m looking at alternatives for another platform. I would be awesome if you could point me in the direction of a good platform.
FrankExign
[url=https://latestcasinobonuses.me/]Участие в турнире – замечательный способ стать обладателем части общего призового фонда, который предоставляется казино или формируется за счет участников. Многие азартные ресурсы предлагают такой тип соревнований на регулярной основе или посвящая определенным событиям. ЛатестКазиноБонус собирает информацию о последних турнирах в онлайн казино, что достойны особого внимания и выкладывает ее сюда. Посты о турнирах содержат данные: какое казино проводит гонку, тип турнира, правила и период состязания, условия участия, а самое главное – размер призового фонда. Турниры отличаются масштабностью главных призов, а для участия в некоторых не нужно особых вложений. Также в таком конкурсе может быть задействован слот, который вам нравится и тогда присоединиться к турниру вполне разумно, ведь к шансам на выигрыш, что дает пейаут, прибавляется еще возможность занять призовое место в турнирной таблице и получить дополнительный приз в реальных деньгах, бонусных средствах или фриспинами.[/url]
GedInsof
eat creampie
https://honda72.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories http://polystar.com.ua/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
StevenBexia
Hello there! [url=http://stromectolxlp.com/]where can i buy stromectol[/url] great web site http://stromectolxlp.com
StevenBexia
Hello! [url=http://stromectolxlp.com/]buy stromectol for humans[/url] great web page http://stromectolxlp.com
FrankExign
[url=https://vetclinika.moscow/]СЕТЬ КРУГЛОСУТОЧНЫХ ВЕТЕРИНАРНЫХ КЛИНИК VETGLOBAL Врачи ветклиники осуществляют весь спектр услуг – от инструментальной и лабораторной диагностики до терапии, хирургии и ведения беременности. Применение рентгеновского оборудования, аппаратов УЗИ и ЭКГ, лапароскопических инструментов обеспечивает безопасность и эффективность лечения. Ветеринары работают круглосуточно и при необходимости выезжают на дом, чтобы исключить ненужный стресс для питомца.[/url]
[url=https://vetclinika.moscow/]КАКИЕ ВИДЫ ОБСЛЕДОВАНИЙ МЫ ПРОВОДИМ НА ДОМУ? Перед госпитализацией следует провести предварительный осмотр питомца и взять анализы, чтобы понять, насколько она необходима. Мы выполняем различные виды диагностики в домашней среде:[/url]
Charmain
Askіng questions are actսally gоod thing if yoս are not understanding аnything entirely, bᥙt tһis post ρresents goߋd understanding еven.
JamesMut
Hello there! [url=http://isotretinoinxp.online/]purchase accutane online no prescription[/url] great internet site http://isotretinoinxp.online
Susanne
Многим представителям сильного пола нужна возможность найти обученную шлюху для незабываемого свидания.
Изучите знаменитый онлайн-портал https://b-n-w.ru, который предназначен ради этой цели! На его страницах находится регулярно обновляемая подборка анкет, которые допустимо просматривать без регистрации. Каждая девушка написала свой рабочий номер, а также опубликовала целую порцию эротических фотографий в отличном качестве!
Насос hag
Для чего нужен в системе отопления циркуляционный насос. Ядро распределение насоса – обеспечить равномерное равно постоянное яппи тепла по общества некоммуникабельной отопительной общественный порядок через котла по и стар и млад периметру строения. НА некоторых способ организации оборотные насосы не нужны. Когда вы выкраивайтесь на формации принятия постановления, как остановить выбор оборотный эрлифт для отопления свой в доску жилища, учитывайте, что энергоресурс насоса кот «сырым» ротором выше, спирт жуть нищенствует в систематическом обслуживании. Данный эрлифт актуален для систем отопления, водоснабжения и вентиляции. В самостоятельной государственное устройство отопления за беспрестанное яппи теплоносителя маленький предопределенною стремительностью отвечает кругообразный насос. Однако этто не один-единственная этиология, числом каковой приобретается это устройство. https://ekavkaz.ucoz.ru/forum/2-18-1 https://fduswat.org.pk/2019/07/05/hello-world/#comment-68222
GedInsof
gangbang creampie anal
http://obs.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://rusfit.club/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
JamesMut
Howdy! [url=http://isotretinoinxp.top/]accutane online[/url] great internet site http://isotretinoinxp.online
FrankExign
[url=https://vetclinika.moscow/]СЕТЬ КРУГЛОСУТОЧНЫХ ВЕТЕРИНАРНЫХ КЛИНИК VETGLOBAL Врачи ветклиники осуществляют весь спектр услуг – от инструментальной и лабораторной диагностики до терапии, хирургии и ведения беременности. Применение рентгеновского оборудования, аппаратов УЗИ и ЭКГ, лапароскопических инструментов обеспечивает безопасность и эффективность лечения. Ветеринары работают круглосуточно и при необходимости выезжают на дом, чтобы исключить ненужный стресс для питомца.[/url]
[url=https://vetclinika.moscow/]КАКИЕ ВИДЫ ОБСЛЕДОВАНИЙ МЫ ПРОВОДИМ НА ДОМУ? Перед госпитализацией следует провести предварительный осмотр питомца и взять анализы, чтобы понять, насколько она необходима. Мы выполняем различные виды диагностики в домашней среде:[/url]
GedInsof
milf cock suck
http://mashinarius.ru/bitrix/redirect.php?event1=&event2=&event3=&goto=https://tubesweet.xyz/categories http://floraland.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
Colby
Hi there, I enjoy reading through your article post. I wanted to write a little comment to support you.
GedInsof
milf black porno
http://mgv-balans.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories https://honda72.ru/bitrix/redirect.php?event1=&event2=&event3=&goto=https://tubesweet.xyz/categories
GedInsof
homemade porn and teens
http://bildj-v.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://ladyhealth.com.ua/bitrix/rk.php?goto=https://tubesweet.xyz/categories
Ronaldloals
Howdy! [url=http://molnupiravir.site/]can i buy Molnupiravir over the counter uk 2022[/url] good site http://molnupiravir.site
GedInsof
big tits anal com
http://itbestseller.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories http://xn–80ahtmekk.xn–p1ai/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories
Ronaldloals
Hi! [url=http://molnupiravir.site/]where can i buy Movfor over the counter uk[/url] good web page http://molnupiravir.site
trust pharmacy canada
canadian medicine https://canadianpharmaciesshop.com/ [url=https://canadianpharmaciesshop.com/]best online pharmacies reviews[/url]
Clement
Viggo Mortensen and Joel Edgerton co-star in the film primarily based on the 2018 rescue of 12 boys and their soccer coach from a flooded cave in Thailand.
Raphael
Colin Farrell and Viggo Mortensen portray real-life rescue divers John Volanthen and Richard Stanton, respectively, with Joel Edgerton, Tom Bateman, and Paul Gleeson additionally starring.
Kaylene
“Thirteen Lives,” penned by William Nicholson, will premiere in theaters for one week beginning July 29 earlier than launching globally on Prime Video August 5.
Hester
I’d like to thank you for the efforts you have put in writing this website. I really hope to view the same high-grade content by you later on as well. In truth, your creative writing abilities has encouraged me to get my very own blog now ;)
Dell
Hello, I enjoy reading through your post. I like to write a little comment to support you.
Irving
Can I just say what a relief to uncover someone who truly knows what they are talking about over the internet. You actually know how to bring an issue to light and make it important. More people need to check this out and understand this side of your story. It’s surprising you’re not more popular since you most certainly have the gift.
Patsy
When I initially commented I clicked the “Notify me when new comments are added” checkbox and now each time a comment is added I get three emails with the same comment.
Is there any way you can remove me from that service? Thanks a lot!
Ashleigh
Hi! Quick question that’s entirely off topic. Do you know how to make your site mobile friendly? My weblog looks weird when viewing from my iphone4. I’m trying to find a theme or plugin that might be able to correct this problem. If you have any suggestions, please share.
Thank you!
Mari
Randi
Excellent post. I certainly love this website. Continue the good work!
Ronaldloals
Howdy! [url=http://molnupiravir.site/]can you buy Molnupiravir over the counter in nz[/url] great web page http://molnupiravir.site
GedInsof
milf 1 on 1
http://belslonik.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories http://oktlife.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories
Дорожные
[url=https://znaki-dorozhnogo-dvizhenija.ru/]Знаки дорожного движения[/url]
Сейчас в течение мегаполисе останавливается шиздец чище и больше экстрим-спорт, то-то патологии правил ща немерено редкость. Для регулирования течения каров а также сжатия численности ДТП производятся приметы, коие значительно упрощают навигацию сверху дороги. <a href=https://znaki-dorozhnogo-dvizhenija.ru/>Знаки дорожного движения</a>
Geneva
WOW just what I was looking for. Ꮯame here by searching fοr where cаn i get generic flagyl pills
Ronaldloals
Hi! [url=http://molnupiravir.site/]how can i buy Molnupiravir[/url] excellent site http://molnupiravir.site
GedInsof
milf hd pov
http://troitskiy-istochnik.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories http://tvgmu.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories
Clark
I’ve been surfing on-line more than three hours as of late, but I never discovered any attention-grabbing article like yours. It’s beautiful worth enough for me. In my view, if all site owners and bloggers made good content material as you probably did, the net will be much more helpful than ever before.
UltraPron
https://www.ultrapron.com/videos/who-is-his-queen-she-deepthroats-bbc-easily/
GedInsof
free and amateur
http://grandtechauto.ru/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories http://vitranet24.com/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories
Ronaldloals
Hi! [url=http://molnupiravir.site/]buy Movfor pills[/url] great internet site http://molnupiravir.site
GedInsof
teens big ass anal
http://2retail.ru/bitrix/rk.php?goto=https://tubesweet.com http://scanpoint.ru/bitrix/click.php?anything=here&goto=https://tubesweet.com
Потолок
[url=https://natjazhnye-potolki-cena-ukraina.dp.ua/]бесщелевой натяжной потолок[/url] [url=https://natjazhnye-potolki-cena-ukraina.dp.ua/]парящий потолок[/url] [url=https://natjazhnye-potolki-cena-ukraina.dp.ua/]натяжной потолок сатин[/url] [url=https://natjazhnye-potolki-cena-ukraina.dp.ua/]натяжной потолок цена[/url] [url=https://natjazhnye-potolki-cena-ukraina.dp.ua/]натяжной потолок в ванной[/url]
Установка современных натяжных потолков мастерами нашей компании — языком не ворочает фотоспособ стильно расширить интерьер помещения вместе с малочисленными расходами. <a href=https://natjazhnye-potolki-cena-ukraina.dp.ua/>глянцевый натяжной потолок</a> <a href=https://natjazhnye-potolki-cena-ukraina.dp.ua/>потолок в ванной</a> <a href=https://natjazhnye-potolki-cena-ukraina.dp.ua/>теневой натяжной потолок</a> <a href=https://natjazhnye-potolki-cena-ukraina.dp.ua/>бесщелевой натяжной потолок</a> <a href=https://natjazhnye-potolki-cena-ukraina.dp.ua/>натяжной потолок в спальне</a>
Ronaldloals
Hi! [url=http://molnupiravir.site/]can i buy Movfor over the counter in uk[/url] good website http://molnupiravir.site
Tahlia
The final half exhibits all of the instances a test has been run towards your credit score report, both because you applied for a mortgage or because a service provider or employer initiated the examine.
Take a look at all of the gastronomical action in this premiere of Planet Green’s latest show, Future Food. So if you enroll to find out what sitcom star you most establish with, the makers of that poll now have access to your personal information. And they have a easy surface for attaching photos, gemstones, fabric or no matter else suits your fancy. Most Web sites that comprise safe personal info require a password also have at the least one password hint in case you overlook. Why do people share embarrassing info on-line? That’s why most on-line scheduling methods permit businesses to create time buckets. Also, RISC chips are superscalar – they will carry out multiple directions at the identical time. But two or three relatively cheap displays can make a world of difference in your computing expertise.
But DVRs have two main flaws – you have to pay for the privilege of utilizing one, and you’re stuck with whatever capabilities the DVR you purchase occurs to include.
GedInsof
teens lesbian porn
http://smi58.ru/bitrix/click.php?anything=here&goto=https://tubesweet.com http://pos-id.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.com
GedInsof
teens fuck there dads
http://sibmotor.su/bitrix/click.php?anything=here&goto=https://tubesweet.com http://md.grouphe.com/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.com
Ronaldloals
Hello there! [url=http://molnupiravir.site/]buy Molnupiravir syrup online[/url] excellent web page http://molnupiravir.site
Rodrick
I do accept as true with all of the ideas you’ve introduced in your post. They are really convincing and will definitely work. Still, the posts are very short for newbies. Could you please lengthen them a bit from next time? Thanks for the post.
Rosie
I’ve been surfing on-line greater than three hours these days, but I never found any attention-grabbing article like yours. It is pretty worth sufficient for me. Personally, if all site owners and bloggers made excellent content as you probably did, the internet can be much more helpful than ever before.
Tawnya
Hey there just wanted to give you a brief heads up and let you know a few of the images aren’t loading correctly. I’m not sure why but I think its a linking issue. I’ve tried it in two different internet browsers and both show the same outcome.
Bret
Hi there colleagues, how is everything, and what you desire to say on the topic of this piece of writing, in my view its truly remarkable for me.
Lane
Howdy, i read your blog from time to time and i own a similar one and i was just curious if you get a lot of spam responses? If so how do you protect against it, any plugin or anything you can advise?
I get so much lately it’s driving me insane so any assistance is very much appreciated.
Debora
I’ve been surfing online more than 3 hours these days, but I by no means found any interesting article like yours. It’s beautiful worth sufficient for me. Personally, if all site owners and bloggers made good content as you probably did, the internet will probably be a lot more helpful than ever before.
Mckenzie
Your style is unique in comparison to other folks I have read stuff from.
I appreciate you for posting when you’ve got the opportunity, Guess I will just book mark this site.
Cecil
Howdy! I know this is kind of off topic but I was wondering which blog platform are you using for this website? I’m getting sick and tired of Wordpress because I’ve had problems with hackers and I’m looking at options for another platform. I would be fantastic if you could point me in the direction of a good platform.
Ofelia
We’re a group of volunteers and opening a new scheme in our community. Your web site offered us with valuable information to work on. You’ve done an impressive job and our whole community will be grateful to you.
JefferyLoalp
Hi! [url=https://buyeddrugx.com/]buy ed pills[/url] good web site https://buyeddrugx.shop
JefferyLoalp
Hi! [url=https://buyeddrugx.com/]cheap erectile dysfunction pills online[/url] very good internet site https://buyeddrugx.com
Leonore
Аwesome! Its truⅼy remarkable ρaragraph, I have got much clear idea conceгning frοm this pаrɑgraph.
JefferyLoalp
Hi! [url=https://buyeddrugx.com/]buy ed pills[/url] good internet site https://buyeddrugx.com
Marshall
Amazing issues here. I’m very glad to see your post. Thanks a lot and I am taking a look ahead to contact you. Will you kindly drop me a mail?
GedInsof
creampies gangbang hd
http://bookhouse.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://mosgorbike.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
JefferyLoalp
Howdy! [url=https://buyeddrugx.com/]erectile dysfunction medicines[/url] great site https://buyeddrugx.com
Korenellykemia
Распродажа формы всех клубов и аксессуаров для мужчин, женщин и детей. Оплата после примерки, купить форму Тоттенхэм 2019 2020. Быстрая и бесплатная доставка по всем городам России. [url=https://footballnaya-forma-tottenxam.ru]форма фк Тоттенхэм в Москве[/url] форма Тоттенхэм 2022 2023 в Москве - [url=http://www.footballnaya-forma-tottenxam.ru]https://footballnaya-forma-tottenxam.ru[/url] [url=http://mx2.radiant.net/Redirect/footballnaya-forma-tottenxam.ru]http://google.am/url?q=http://footballnaya-forma-tottenxam.ru[/url]
[url=https://gctswabi.com/2022/01/18/mid-term-examination-2022/#comment-1251]Спортивная одежда для футбола с примеркой перед покупкой и быстрой доставкой в любой город РФ.[/url] b7ab000
GedInsof
slutty teens
http://school595.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://fabrica-aztec.com/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories
JefferyLoalp
Hello there! [url=https://buyeddrugx.com/]erection pills viagra online[/url] beneficial website https://buyeddrugx.com
Kennith
Wow that was odd. I just wrote an really long comment but after I clicked submit my comment didn’t show up. Grrrr…
well I’m not writing all that over again. Regardless, just wanted to say fantastic blog!
Gladis
I will immediately seize your rss as I can not in finding your email subscription hyperlink or e-newsletter service. Do you’ve any? Kindly allow me understand in order that I could subscribe. Thanks.
Arlie
Hello this is somewhat of off topic but I was wanting to know if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding experience so I wanted to get advice from someone with experience. Any help would be greatly appreciated!
Roxie
Hey there, You’ve done a great job. I will certainly digg it and personally suggest to my friends. I am sure they will be benefited from this site.
Katharina
Hey there, You have done a fantastic job. I’ll certainly digg it and personally recommend to my friends.
I’m sure they will be benefited from this site.
Rashad
If you would like to increase your knowledge simply keep visiting this web page and be updated with the latest news update posted here.
Marietta
Thank you for sharing your info. I truly appreciate your efforts and I will be waiting for your further post thanks once again.
sosnitikc
[url=https://sosnitikc.dp.ua/]sosnitikc[/url]
Pin Up - вход а также фоторегистрация язык легального букмекера Номер Ап ру. Экспресс-обзор официознного сайта. Целиком экспресс-информация что касается букмекерской фирме Пин Уп <a href=https://sosnitikc.dp.ua/>sosnitikc</a>
JeffreyKaK
Hello there! [url=https://edpill.shop/]ed medications[/url] great web site https://edpill.shop
GedInsof
sexy teens models
http://vlmj.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://rmsft.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
GedInsof
ass anal milf
http://metplus-vrn.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://alta-energo.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories
JeffreyKaK
Hello there! [url=https://edpill.shop/]erectile dysfunction medications[/url] excellent web site https://edpill.shop
Kristina
Touche. Solid arguments. Keep up the great spirit.
GedInsof
milf игры i
http://macbookhelp.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://it-hive.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
Josefina
Нi my friend! I ԝant to say that this post is amazing, great written and comе with approⲭimately all vital infos. І’d like to peer extra posts ⅼike this .
Luz
Quality articles is the crucial to interest the viewers to go to see the web site, that’s what this website is providing.
Ialeflorne
Shoplyfters Harmony Wonder and Elle Vonevas hairy pussy gets romped hard! https://wankmovie.com/daily/cassidy-blanc-xxx-movie.html She met this tranny, Patricia Kimberly, and went back to her hotel with her
GedInsof
gangbang black
http://bsme.moscow/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories http://dzvr.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories
Josh
Truly no matter if someone doesn’t understand afterward its up to other visitors that they will assist, so here it takes place.
Tyrell
I’m not sure where you’re getting your information, but good topic. I needs to spend some time learning much more or understanding more. Thanks for fantastic info I was looking for this information for my mission.
Muhammad
Hey there, You’ve done a fantastic job. I will definitely digg it and personally suggest to my friends. I’m confident they’ll be benefited from this site.
Kirsten
Very nice post. I just stumbled upon your weblog and wanted to say that I have truly enjoyed browsing your blog posts. In any case I will be subscribing to your feed and I hope you write again very soon!
Kazuko
My coder is trying to persuade me to move to .net from PHP. I have always disliked the idea because of the expenses. But he’s tryiong none the less. I’ve been using Movable-type on various websites for about a year and am concerned about switching to another platform. I have heard fantastic things about blogengine.net. Is there a way I can transfer all my wordpress content into it? Any help would be really appreciated!
Lorrie
I go to see day-to-day some websites and websites to read posts, except this blog gives feature based articles.
Doug
Thankfulness to my father who informed me on the topic of this web site, this web site is in fact remarkable.
Erica
Thanks designed for sharing such a nice thinking, post is good, thats why i have read it entirely
GedInsof
all girl anal sex
http://www.technoschock.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://otdelka-market.ru/bitrix/redirect.php?event1=&event2=&event3=&goto=https://tubesweet.xyz/categories
Бухгалтера
[url=https://buhgalterskie-uslugi-gba.ru/]Бухгалтерские услуги[/url]
Проводка счетоводного учета в течение картине книг бухгалтерского учета также проводка налоговой документации, которое складывается из: отдельный учет для цельнее налогообложения продуктов равным образом услуг эскомпт коренных средств равно нематериальных активов вместе один-другой расчетом амортизации подготовка да эйс налоговых деклараций в течение налоговую инспекцию, осведомленную чтобы Поставщика <a href=https://buhgalterskie-uslugi-gba.ru/>Бухгалтерские услуги</a>
GedInsof
milf lust
https://kazan.yitservice.ru/bitrix/redirect.php?event1=&event2=&event3=&goto=https://tubesweet.xyz/categories http://browland.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
Laverne
Hi there to all, for the reason that I am genuinely eager of reading this website’s post to be updated daily. It includes pleasant information.
GedInsof
mistress and cuckold
http://vsg.ua/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories http://snab-a.ru/bitrix/redirect.php?event1=&event2=&event3=&goto=https://tubesweet.xyz/categories
Ralf
hey there and thank you for your information – I have certainly picked up anything new from right here. I did however expertise some technical points using this site, since I experienced to reload the site a lot of times previous to I could get it to load correctly. I had been wondering if your web host is OK? Not that I’m complaining, but sluggish loading instances times will very frequently affect your placement in google and can damage your high quality score if advertising and marketing with Adwords.
Anyway I’m adding this RSS to my email and can look out for a lot more of your respective intriguing content. Ensure that you update this again soon.
Celesta
гостиничные чеки купить спб
GedInsof
milf in bikini
http://gektor-nsk.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://xn–80aaco0acjmv.xn–p1ai/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
Barry
I know this if off topic but I’m looking into starting my own weblog and was curious what all is required to get set up? I’m assuming having a blog like yours would cost a pretty penny?
I’m not very internet savvy so I’m not 100% sure.
Any recommendations or advice would be greatly appreciated. Many thanks
GedInsof
video model teens
http://yaroshenkonv.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://epages.su/bitrix/rk.php?goto=https://tubesweet.xyz/categories
sosnitikd
[url=https://vkontaktemp3.ru/]sosnitikd[/url]
Климатическая техника чтобы комплекса комнат именуется мультисплит-системами. Они мыслят собою привычные блоки внутренней равным образом наружной поправки, <a href=https://vkontaktemp3.ru/>sosnitikd</a>
RichardJouse
Howdy! [url=http://spicyypharm.top/]rx online[/url] very good web site http://spicyypharm.top
Pauladut
Здравствуйте хорошая мысль но Так же хочу добавит
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=U5gBMUfUZWI https://www.youtube.com/watch?v=z6u1sF_Zd_U https://www.youtube.com/watch?v=NeAdaxLj8a8 https://www.youtube.com/watch?v=4gAGRFKp–s https://www.youtube.com/watch?v=O6-r20g4bxk https://www.youtube.com/watch?v=cHzIH5YSpBs https://www.youtube.com/watch?v=pqHIjQNSCLE
[url=https://www.youtube.com/watch?v=U5gBMUfUZWI]How to open 1xbet account in Bangladesh best free bet promo code Bangla and India for registration[/url] [url=https://www.youtube.com/watch?v=z6u1sF_Zd_U]How to open 1xbet account in Bangladesh best free bet promo code Bangla and India for registration[/url] [url=https://www.youtube.com/watch?v=NeAdaxLj8a8]How to open 1xbet account in Bangladesh best free bet promo code Bangla and India for registration[/url] [url=https://www.youtube.com/watch?v=4gAGRFKp–s]How to open 1xbet account in Bangladesh best free bet promo code Bangla and India for registration[/url]
[url=https://www.youtube.com/playlist?list=PLhTL01CCZ4DrxcFQ5aJfpA1D3-VNsM1Kx]1xbet cricket No risk 1xbet loss cover tricks 1xbet prediction Bangla Tutorial online tips Bangla - 1xbet telegram channel promo code bangla[/url] [url=https://www.youtube.com/playlist?list=PLPH-IC8r4kFEGrtyEkWRGqUnLrjAeHCUH] How to withdraw 1xbet Bonus Bangla 2022 - 1X BET promo code Bangladesh 2022[/url] [url=https://www.youtube.com/playlist?list=PLF_zC7NDnQf-EW9AtuKx7RCZRzF2Bpwej]1xbet Bangladesh 2022 1xbet promo code Bangladesh 1xbet promo code Bangladesh 2022[/url] [url=https://www.youtube.com/playlist?list=PLGwWlnPV-nr9IuU8OG4-Zj1WjGlvWUf6J]1XBET bangla tutorial promo code - 1xbet Deposit Problem 2022 1xbet Deposit Rejected 2022 1xbet Deposit Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLgwoHAwRWuSXfShVoou9gnczQ5RzhhZiI]How to open 1XBET account in bangladesh - 1xbet a to z bangla tutorial 2022.how to open 1xbet account bangla.how to create 1xbet account bd[/url]
https://www.youtube.com/playlist?list=PLzS_Kdj5GfYpI_dEgfbNw9r53t4QsBs4u https://www.youtube.com/playlist?list=PLgQud9YXHpBV0JYFvslICUISV5LUgrdGH https://www.youtube.com/playlist?list=PLzm4MUwMiXL8yP9YfYh9ec9fRqAYd464_ https://www.youtube.com/playlist?list=PLGDCQlhdwIRCdfUJ9IKK4I3JzlfV8oYP8
RX675HGKFjgf9025
GedInsof
teens and smoking
http://ckr.avto-trakt.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://enzolab.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
RichardJouse
Howdy! [url=http://spicyypharm.top/]online pharmacies[/url] great site http://spicyypharm.top
Umunmovib
https://wankmovie.com/daily/candidking-com.html Sexually Excited Plump GF love fucking and getting cum on her face-P2
Leonore
When I originally commented I clicked the “Notify me when new comments are added” checkbox and now each time a comment is added I get four e-mails with the same comment. Is there any way you can remove people from that service?
Many thanks!
RichardJouse
Hello there! [url=http://spicyypharm.top/]canadian xanax[/url] great web page http://spicyypharm.top
GedInsof
creampie compilation pussy
http://ooomsv.ru/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories http://www.innov-rosatom.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
Lorene
What’s Going down i’m new to this, I stumbled upon this I have discovered It positively helpful and it has aided me out loads. I am hoping to give a contribution & help other customers like its helped me. Good job.
Leonida
Definitely believe that which you stated. Your favourite justification seemed to be on the net the simplest thing to understand of. I say to you, I certainly get irked at the same time as people consider worries that they just don’t understand about. You controlled to hit the nail upon the top as well as outlined out the whole thing with no need side-effects , folks could take a signal. Will probably be back to get more. Thank you
Caleb
Hi there, its good paragraph about media print, we all be familiar with media is a fantastic source of data.
Sheldon
Do you mind if I quote a few of your posts as long as I provide credit and sources back to your webpage? My website is in the exact same niche as yours and my users would truly benefit from a lot of the information you provide here. Please let me know if this okay with you. Thanks a lot!
GedInsof
creampie peeing
http://europroject.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://mc-camp.ru/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories
Don
For newest information you have to pay a quick visit the web and on internet I found this web site as a most excellent website for newest updates.
Aracelis
I really love your website.. Great colors & theme. Did you build this amazing site yourself? Please reply back as I’m wanting to create my very own site and would like to know where you got this from or just what the theme is named. Many thanks!
Julienne
Howdy very cool blog!! Man .. Excellent .. Wonderful .. I’ll bookmark your web site and take the feeds also? I am happy to find numerous helpful information here in the submit, we need develop extra strategies on this regard, thanks for sharing. . . . . .
Delbert
Thanks very interesting blog!
Jeff
Thanks , I’ve just been looking for info about this subject for a while and yours is the best I have came upon till now. However, what concerning the bottom line? Are you certain concerning the source?
Natisha
I don’t even understand how I ended up right here, but I assumed this publish used to be great. I do not know who you might be however certainly you are going to a well-known blogger if you are not already.
Cheers!
Pamela
Hi! Do you know if they make any plugins to protect against hackers?
I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
Andre
Superb, what a website it is! This webpage gives useful data to us, keep it up.
GedInsof
milf on twitter
http://school571spb.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://xn–d1abdw2b.net/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
GedInsof
links for teens
http://www.aquainterio.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://kristall43.ru/bitrix/rk.php?goto=https://tubesweet.xyz/categories
Emunmovib
Busty milf Yasmin Scott teaches sex Cali Sparks and her boyfriend https://wankmovie.com/daily/naughty-nanny.html Shes eager to feel a thick, hard cock splitting and stretching her pussy and nows her chance to ride the big bone, having all kinds of intense orgasms and showing him a
GedInsof
porn anal big tits
http://cosmeticshop.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://xn—-8sbec4aclrbhbccnwjr2k.xn–p1ai/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
GedInsof
hot teens pussies
http://gklink.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories http://protennis.by/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
Jamila
прописка в Москве
GedInsof
vk com teens
http://principcomp.ru/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories http://1c-rating.com/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
GedInsof
anal fuck video
http://kolesatyt.ru/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories http://inostezia.su/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
Carley
Good post. I learn something new and challenging on websites I stumbleupon everyday. It’s always useful to read through content from other writers and practice something from other websites.
Kara
Resumes will be accepted and reviewed straight away and continue until the position is filled.
GedInsof
milf 35
http://shuttle-logistic.pro/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://zapravka39.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
GedInsof
big ass amateurs
http://irkutsk.vape.academy/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://www.kayaker.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
Albertina
Hi there, its good post regarding media print, we all be aware of media is a wonderful source of facts.
Mickie
Remarkable! Its genuinely awesome post, I have got much clear idea concerning from this post.
Anya
I every time emailed this web site post page to all my friends, for the reason that if like to read it afterward my links will too.
Domingo
Hmm it appears like your site ate my first comment (it was extremely long) so I guess I’ll just sum it up what I wrote and say, I’m thoroughly enjoying your blog. I too am an aspiring blog blogger but I’m still new to everything.
Do you have any suggestions for rookie blog writers? I’d genuinely appreciate it.
Arthur
You should be a part of a contest for one of the best sites on the internet. I will recommend this site!
Lloyd
Excellent web site you’ve got here.. It’s hard to find high quality writing like yours nowadays. I honestly appreciate people like you! Take care!!
Lino
When someone writes an post he/she maintains the image of a user in his/her mind that how a user can know it. So that’s why this post is perfect. Thanks!
GedInsof
in anal photos
http://milazlata.ru/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories https://sk-tmk.ru/bitrix/redirect.php?event1=&event2=&event3=&goto=https://tubesweet.xyz/categories
Amelia
Simply desire to say your article is as surprising. The clearness to your put up is just cool and that i can suppose you’re knowledgeable on this subject. Well together with your permission allow me to clutch your RSS feed to stay updated with coming near near post. Thanks 1,000,000 and please keep up the gratifying work.
KendraEmurf
Салют Поддерживаю И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=cnFb5JF9I74 https://www.youtube.com/watch?v=Yo9WounjURA https://www.youtube.com/watch?v=2VibXiXfzCo https://www.youtube.com/watch?v=a4K_s3jd5sA https://www.youtube.com/watch?v=Ooe7Zhr8Ios https://www.youtube.com/watch?v=nAUbLZOMfU8 https://www.youtube.com/watch?v=MJhG3LuCWTw
https://www.youtube.com/playlist?list=PLNzG2x9bf1wxkJXXWFQrI2Qq-8VHXgD1u https://www.youtube.com/playlist?list=PLNzG2x9bf1wwRwGZrF1GAY771VcANiV7y https://www.youtube.com/playlist?list=PLNzG2x9bf1wwP_ovG_-uF6hlQU-mQiZ2d https://www.youtube.com/playlist?list=PLNzG2x9bf1wy-k1qXGGu6URJzKrQC_QHN https://www.youtube.com/playlist?list=PLNzG2x9bf1wzdbxfnsIl9CKPPnK18JbXc https://www.youtube.com/playlist?list=PLNzG2x9bf1wxlt3We38spiaBkzJIxbnDI https://www.youtube.com/playlist?list=PLNzG2x9bf1wytmSbtsBC80Wyx1FrWF8d1 https://www.youtube.com/playlist?list=PLNzG2x9bf1wwHUZ0Ddknog6XlcoFTdN1j https://www.youtube.com/playlist?list=PLNzG2x9bf1wxLEtKqc8b0aEkdmmtcPAK8
KendraEmurf
Салют со свем согласен и даже более И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=ZGiZvXquPcQ https://www.youtube.com/watch?v=g_d0Zyl2VqE https://www.youtube.com/watch?v=vIZD9oOl0tg https://www.youtube.com/watch?v=HLrq-tjYTx8 https://www.youtube.com/watch?v=TMBWt3Xf5xw https://www.youtube.com/watch?v=fieWXlMi01o https://www.youtube.com/watch?v=GhBNvgzg5oc
https://www.youtube.com/playlist?list=PLneJ0QN_YNk5ULpCgAhMN9U3v81wiIn8S https://www.youtube.com/playlist?list=PLneJ0QN_YNk41flytvb9sWU5JTs6hcxfP https://www.youtube.com/playlist?list=PLneJ0QN_YNk4Zusz1_1dgbXeXgseMEOSO https://www.youtube.com/playlist?list=PLneJ0QN_YNk6rxCx20U_NywIFLeEfFHPx https://www.youtube.com/playlist?list=PLneJ0QN_YNk6gyggk4NIwX_5_bBgt7gLG https://www.youtube.com/playlist?list=PLneJ0QN_YNk4ZSS9rz5-KjdPEMCh1gK-W https://www.youtube.com/playlist?list=PLneJ0QN_YNk60vQU3WjmR51gB2h3o7XCw https://www.youtube.com/playlist?list=PLneJ0QN_YNk5vQGg3wjCLGYZZYjkcnKOB https://www.youtube.com/playlist?list=PLneJ0QN_YNk6kF0CYJX1TkYF7bVbWY6hl
KendraEmurf
Привет Поддерживаю Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
https://www.youtube.com/playlist?list=PLneJ0QN_YNk6baz9zeudBhUCzYlGtPE_0 https://www.youtube.com/playlist?list=PLneJ0QN_YNk7Y8tx-VgSnl7ubzdIKCRnX https://www.youtube.com/playlist?list=PLneJ0QN_YNk5iRlC3MVh2RsAFmB88YSZE https://www.youtube.com/playlist?list=PLneJ0QN_YNk6OLDy8SaIvgWsMxEHQubOo https://www.youtube.com/playlist?list=PLneJ0QN_YNk7jFysr32Sf6Q5yu9B7eoH2 https://www.youtube.com/playlist?list=PLneJ0QN_YNk5ThXvfx3JOuKX-omxqpcHK https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBrBmHmAX_RHanGvmD464yf https://www.youtube.com/playlist?list=PLRcnUiG5ZvBAUZm9GihHfYfDK_Ftm7HoM
KendraEmurf
Здравствуйте хорошая мысль но И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=cnFb5JF9I74 https://www.youtube.com/watch?v=Yo9WounjURA https://www.youtube.com/watch?v=2VibXiXfzCo https://www.youtube.com/watch?v=a4K_s3jd5sA https://www.youtube.com/watch?v=Ooe7Zhr8Ios https://www.youtube.com/watch?v=nAUbLZOMfU8 https://www.youtube.com/watch?v=MJhG3LuCWTw
https://www.youtube.com/playlist?list=PLneJ0QN_YNk5ULpCgAhMN9U3v81wiIn8S https://www.youtube.com/playlist?list=PLneJ0QN_YNk41flytvb9sWU5JTs6hcxfP https://www.youtube.com/playlist?list=PLneJ0QN_YNk4Zusz1_1dgbXeXgseMEOSO https://www.youtube.com/playlist?list=PLneJ0QN_YNk6rxCx20U_NywIFLeEfFHPx https://www.youtube.com/playlist?list=PLneJ0QN_YNk6gyggk4NIwX_5_bBgt7gLG https://www.youtube.com/playlist?list=PLneJ0QN_YNk4ZSS9rz5-KjdPEMCh1gK-W https://www.youtube.com/playlist?list=PLneJ0QN_YNk60vQU3WjmR51gB2h3o7XCw https://www.youtube.com/playlist?list=PLneJ0QN_YNk5vQGg3wjCLGYZZYjkcnKOB https://www.youtube.com/playlist?list=PLneJ0QN_YNk6kF0CYJX1TkYF7bVbWY6hl
GedInsof
teens daughter fuck
http://xn–80aaxnbbbh6h3a.xn–p1ai/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://bitrixhost.com/bitrix/click.php?anything=here&goto=https://tubesweet.xyz/categories
BrendaTed
Hi согласен со всем Так же хочу добавить Parimatch зеркало сайта работающее последнее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=Sn4WZyanEh4 https://www.youtube.com/watch?v=Z_GAElA0QKc https://www.youtube.com/watch?v=1uFOC48XHG8 https://www.youtube.com/watch?v=k6hfH6RB9V0 https://www.youtube.com/watch?v=P4cadSLQ1rU https://www.youtube.com/watch?v=p2xEzaH0FRo https://www.youtube.com/watch?v=AROs8UqEujI https://www.youtube.com/watch?v=OEpFTKWUyY8 https://www.youtube.com/watch?v=9QYSZmWmRJY https://www.youtube.com/watch?v=TvkP3CrEJic https://www.youtube.com/watch?v=mi0z1KvoZ2U
[url=https://www.youtube.com/watch?v=Sn4WZyanEh4]ПРОМОКОД Париматч фриспины Parimatch на бесплатные фриспины за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=1uFOC48XHG8]ПРОМОКОД Париматч фриспины Parimatch на бесплатные фриспины за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=Z_GAElA0QKc]ПРОМОКОД Париматч фриспины Parimatch на бесплатные фриспины за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=k6hfH6RB9V0]Промокод на фриспины 77 Париматч с депозитом только за регистрацию 2022 Parimatch[/url] [url=https://www.youtube.com/watch?v=P4cadSLQ1rU]Париматч Украина игровые автоматы бонус за регистрацию бездепозитный на реальные деньги[/url] [url=https://www.youtube.com/watch?v=p2xEzaH0FRo]Париматч игровые автоматы минимальный депозит їх бонуси за регистрацию отзывы[/url]
https://www.youtube.com/playlist?list=PLnsnFM60Rk1S_Jargkz9EpKhVqjn6itat https://www.youtube.com/playlist?list=PLoV15PAdIrbtRnmM5b2UJt31V7wXVZ7G9 https://www.youtube.com/playlist?list=PLubYmKbhMVAg4jncddzEkbOMHLUf_GccA https://www.youtube.com/playlist?list=PL1R7UsD2cHeCdVrhJ6TOER4bxWQ7ApY5i https://www.youtube.com/playlist?list=PLlh1XFwgra_2pLsVUhhfouib0cwDVgb4y https://www.youtube.com/playlist?list=PLiHyRoZH_meBNtosHJRoHt6B7p1S2RiFS https://www.youtube.com/playlist?list=PLCsGp5QHVXdo58bY_AH_TZxNNqpvK0aGC https://www.youtube.com/playlist?list=PLlh1XFwgra_2pLsVUhhfouib0cwDVgb4y
https://www.youtube.com/playlist?list=PL1D6Led19hjDG61mPEC8Wii-AKMHOCtfO https://www.youtube.com/playlist?list=PLBLO37Hy6RNAUMgYwQErjaDuI-EAlDfdE https://www.youtube.com/playlist?list=PLN5HOHzLUbahEWq8YAM8WhD-4Y58ANPp- https://www.youtube.com/playlist?list=PL90A0HJl0Q_VSkHt71-0_JYKPU4CDYCqL https://www.youtube.com/playlist?list=PLtuKjXLNTK35Hk1mJ0DnsBS9W2wkm5TXe https://www.youtube.com/playlist?list=PLYU7T1QT5d_P04Rc4csQNgOq4XZn2J3UQ https://www.youtube.com/playlist?list=PLZNC0fOGfVEi2ncf0aX9f-DbSDIkF-lYi https://www.youtube.com/playlist?list=PLvwjeYnP3Hn5hz9bbijf_DeHqWFJiZ-2K
https://vk.com/parimatch_poker_otzyvy_igrokov
[url=https://vk.com/parimatch_zerkalo_rabohcee_2021]Париматч зеркало последнее рабочее 2021[/url]
Christena
Don’t wait for a small crack to become an emergency glass repair problem.
Maryellen
What’s Happening i’m new to this, I stumbled upon this I’ve found It absolutely helpful and it has aided me out loads. I’m hoping to give a contribution & aid other customers like its helped me. Great job.
GedInsof
videos cuckold femdom
http://techwiregroup.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://repsol.by/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories
BrendaTed
Привет Поддерживаю Так же хочу добавить Parimatch зеркало последнее работающее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=OEpFTKWUyY8 https://www.youtube.com/watch?v=9QYSZmWmRJY https://www.youtube.com/watch?v=TvkP3CrEJic https://www.youtube.com/watch?v=mi0z1KvoZ2U
[url=https://www.youtube.com/watch?v=k6hfH6RB9V0]Промокод на фриспины 77 Париматч с депозитом только за регистрацию 2022 Parimatch[/url] [url=https://www.youtube.com/watch?v=P4cadSLQ1rU]Париматч Украина игровые автоматы бонус за регистрацию бездепозитный на реальные деньги[/url] [url=https://www.youtube.com/watch?v=p2xEzaH0FRo]Париматч игровые автоматы минимальный депозит їх бонуси за регистрацию отзывы[/url] [url=https://www.youtube.com/watch?v=AROs8UqEujI]Париматч промокод при регистрации без депозита 2022 promo bonus Parimatch за верификацию[/url]
https://www.youtube.com/playlist?list=PLw09JdsboSXeVRhQhElN9Rs19lrlwpEIr https://www.youtube.com/playlist?list=PLTivr89Sv4_1PUD7y4uv2Q6juTvOJutqe https://www.youtube.com/playlist?list=PLV9fx-snYv-BcNVbkklhgBK39od15FTCs https://www.youtube.com/playlist?list=PL2xeIlkrOHgCac9hUQA_LLq_vDtPMcs_c https://www.youtube.com/playlist?list=PLmwETSG8roNeOeyJ-2J0oyZv9AvyQCftJ https://www.youtube.com/playlist?list=PLZuLEl52mRuPCSDyuA1MV0J-JTzXdHudK https://www.youtube.com/playlist?list=PLbQMT4K44x55nLUqLm8plNyfycQsRTv1p https://www.youtube.com/playlist?list=PLEuGhPK0FqdczSxTkhFghPr8R46a5BWx3 https://www.youtube.com/playlist?list=PLHegGFkV5quHxhI49_VWim9ahv9D-FFBV
[url=https://www.youtube.com/playlist?list=PL1D6Led19hjDG61mPEC8Wii-AKMHOCtfO]Промокод БОНУС Париматч Слоты Которые Дают 2022 самые выигрышные дающие с бонусами популярные слот в Parimatch[/url] [url=https://www.youtube.com/playlist?list=PLBLO37Hy6RNAUMgYwQErjaDuI-EAlDfdE]Промокод БОНУС Париматч Слоты Которые Дают 2022 самые выигрышные дающие с бонусами популярные слот в Parimatc Украина детальный обзор отзывы[/url] [url=https://www.youtube.com/playlist?list=PLN5HOHzLUbahEWq8YAM8WhD-4Y58ANPp-]Париматч бонус за верификацию - Пари Матч бонусы при регистрации - как забрать бонус в Parimatch[/url] [url=https://www.youtube.com/playlist?list=PL90A0HJl0Q_VSkHt71-0_JYKPU4CDYCqL]Обзор онлайн Париматч контора Parimatch бонус - Промокод Париматч Как получить фриспины и бонусы в Пари Матч без депозита[/url] [url=https://www.youtube.com/playlist?list=PLtuKjXLNTK35Hk1mJ0DnsBS9W2wkm5TXe]Промокод Париматч 2022 Украина - Как зарегистрироваться на Пари-Матч и получить бонус[/url] [url=https://www.youtube.com/playlist?list=PLYU7T1QT5d_P04Rc4csQNgOq4XZn2J3UQ]Где ввести промокод Париматч Беларусь на промокоды Parimatch Украина Пари Матч КЗ 2022[/url] [url=https://www.youtube.com/playlist?list=PLZNC0fOGfVEi2ncf0aX9f-DbSDIkF-lYi]Пари Матч Промокод при регистрации - Обзор казино Париматч Слоты, Бонусы, Регистрация, Вход в Parimatch Casino[/url] [url=https://www.youtube.com/playlist?list=PLvwjeYnP3Hn5hz9bbijf_DeHqWFJiZ-2K]Пари Матч промокод фриспины Обзор Онлайн Казино Париматч Parimatch Букмекерская Контора Париматч Бонус[/url]
https://vk.com/zerkalo_pari_match_rabochee
[url=https://vk.com/parimatch_poker_otzyvy_igrokov]Париматч зеркало сайта вход отзывы игроков 2022[/url]
GedInsof
cuckold in russian
http://intendant.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://dddgazeta.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
Johanna
It’s not my first time to go to see this web site, i am browsing this website dailly and take good data from here every day.
Eula
This piece of writing is actually a nice one it helps new net people, who are wishing in favor of blogging.
Stanton
Hurrah! Finally I got a web site from where I can truly take valuable information regarding my study and knowledge.
Veta
Do you mind if I quote a couple of your articles as long as I provide credit and sources back to your website? My blog site is in the exact same area of interest as yours and my users would genuinely benefit from some of the information you present here.
Please let me know if this ok with you. Cheers!
Marylyn
Fantastic beat ! I would like to apprentice while you amend your web site, how can i subscribe for a blog website? The account aided me a acceptable deal. I had been tiny bit acquainted of this your broadcast provided bright clear concept
Kali
Thanks for one’s marvelous posting! I quite enjoyed reading it, you happen to be a great author. I will remember to bookmark your blog and will often come back someday. I want to encourage continue your great writing, have a nice day!
Lamar
Howdy! Do you use Twitter? I’d like to follow you if that would be okay.
I’m definitely enjoying your blog and look forward to new posts.
GedInsof
porno big tits anal
http://xn–e1akbmnee.xn–p1ai/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://store2.innet-it.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
BrendaTed
Здрасте хорошая мысль но Так же по теме Parimatch зеркало последнее работающее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=GgygicCW120 https://www.youtube.com/watch?v=jI9fRdPcauM https://www.youtube.com/watch?v=r_HRxxhD9oQ https://www.youtube.com/watch?v=PwH_uOJD7P8 https://www.youtube.com/watch?v=CU6eili0m0g https://www.youtube.com/watch?v=oxbcvcGd91w https://www.youtube.com/watch?v=yDfhfQGaKHQ https://www.youtube.com/watch?v=je1OM3vzLPI https://www.youtube.com/watch?v=ZtLOh64g6Zo
[url=https://www.youtube.com/watch?v=CU6eili0m0g]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=oxbcvcGd91w]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=yDfhfQGaKHQ]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=je1OM3vzLPI]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=ZtLOh64g6Zo]Париматч промокод при регистрации без бонус депозита 2022 promo bonus Parimatch за верификацию[/url]
https://www.youtube.com/playlist?list=PLw09JdsboSXeVRhQhElN9Rs19lrlwpEIr https://www.youtube.com/playlist?list=PLTivr89Sv4_1PUD7y4uv2Q6juTvOJutqe https://www.youtube.com/playlist?list=PLV9fx-snYv-BcNVbkklhgBK39od15FTCs https://www.youtube.com/playlist?list=PL2xeIlkrOHgCac9hUQA_LLq_vDtPMcs_c https://www.youtube.com/playlist?list=PLmwETSG8roNeOeyJ-2J0oyZv9AvyQCftJ https://www.youtube.com/playlist?list=PLZuLEl52mRuPCSDyuA1MV0J-JTzXdHudK https://www.youtube.com/playlist?list=PLbQMT4K44x55nLUqLm8plNyfycQsRTv1p https://www.youtube.com/playlist?list=PLEuGhPK0FqdczSxTkhFghPr8R46a5BWx3 https://www.youtube.com/playlist?list=PLHegGFkV5quHxhI49_VWim9ahv9D-FFBV
[url=https://www.youtube.com/playlist?list=PL1D6Led19hjDG61mPEC8Wii-AKMHOCtfO]Промокод БОНУС Париматч Слоты Которые Дают 2022 самые выигрышные дающие с бонусами популярные слот в Parimatch[/url] [url=https://www.youtube.com/playlist?list=PLBLO37Hy6RNAUMgYwQErjaDuI-EAlDfdE]Промокод БОНУС Париматч Слоты Которые Дают 2022 самые выигрышные дающие с бонусами популярные слот в Parimatc Украина детальный обзор отзывы[/url] [url=https://www.youtube.com/playlist?list=PLN5HOHzLUbahEWq8YAM8WhD-4Y58ANPp-]Париматч бонус за верификацию - Пари Матч бонусы при регистрации - как забрать бонус в Parimatch[/url] [url=https://www.youtube.com/playlist?list=PL90A0HJl0Q_VSkHt71-0_JYKPU4CDYCqL]Обзор онлайн Париматч контора Parimatch бонус - Промокод Париматч Как получить фриспины и бонусы в Пари Матч без депозита[/url] [url=https://www.youtube.com/playlist?list=PLtuKjXLNTK35Hk1mJ0DnsBS9W2wkm5TXe]Промокод Париматч 2022 Украина - Как зарегистрироваться на Пари-Матч и получить бонус[/url] [url=https://www.youtube.com/playlist?list=PLYU7T1QT5d_P04Rc4csQNgOq4XZn2J3UQ]Где ввести промокод Париматч Беларусь на промокоды Parimatch Украина Пари Матч КЗ 2022[/url] [url=https://www.youtube.com/playlist?list=PLZNC0fOGfVEi2ncf0aX9f-DbSDIkF-lYi]Пари Матч Промокод при регистрации - Обзор казино Париматч Слоты, Бонусы, Регистрация, Вход в Parimatch Casino[/url] [url=https://www.youtube.com/playlist?list=PLvwjeYnP3Hn5hz9bbijf_DeHqWFJiZ-2K]Пари Матч промокод фриспины Обзор Онлайн Казино Париматч Parimatch Букмекерская Контора Париматч Бонус[/url]
https://vk.com/bk_pari_match_zerkalo_vhod
[url=https://vk.com/bukmekerskaya_kontora_parimatch]Париматч зеркало бк все о букмекерских конторах[/url]
BrendaTed
Hi со свем согласен Так же по теме Parimatch зеркало последнее работающее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=R7C2kb-jR_A https://www.youtube.com/watch?v=FWZqhvCT8Vk https://www.youtube.com/watch?v=qg5QzwGZYdY https://www.youtube.com/watch?v=Fka1Vy61sgg https://www.youtube.com/watch?v=q8FBllok5Zg https://www.youtube.com/watch?v=SDAYlhQL1aQ https://www.youtube.com/watch?v=sBWiZc3uUAw https://www.youtube.com/watch?v=Rt7eLCgnAhQ https://www.youtube.com/watch?v=mPuw5kpYgbk
[url=https://www.youtube.com/watch?v=CU6eili0m0g]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=oxbcvcGd91w]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=yDfhfQGaKHQ]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=je1OM3vzLPI]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=ZtLOh64g6Zo]Париматч промокод при регистрации без бонус депозита 2022 promo bonus Parimatch за верификацию[/url]
https://www.youtube.com/playlist?list=PLgmA4Qx-NO-R4lIJU7Wp5x_RpLAfW7m9M https://www.youtube.com/playlist?list=PLlOP0S2i0zJRMv9A_8exWBE26U0pOetyM https://www.youtube.com/playlist?list=PL-oOWrKcXD00DmJbVd5SbliXo3sJR0mPn https://www.youtube.com/playlist?list=PLlprnaVTmuxLRGNtRWW8UInuNku6eMT6j https://www.youtube.com/playlist?list=PLD8Bx5tfu2pB8o1G2GsdrgTsq8OGxtAEd https://www.youtube.com/playlist?list=PLx1-Xp1bcravZrD9iaeb5St48x5Bv5uhe https://www.youtube.com/playlist?list=PL8XebQPlV77AXeikiiql_fIyBE234KH-B https://www.youtube.com/playlist?list=PLCOaM6CLKyZ861-RCfEX5MTkE4u4R0Wnc https://www.youtube.com/playlist?list=PLP14Wt7nBIGNY-5hikQzMdhg4vBi5Pr8-
https://www.youtube.com/playlist?list=PLW6Elf6cZK1LuxbvdHN4lgTVZruk56Iln https://www.youtube.com/playlist?list=PLqP_K6SSpMSHwzaPnha4AhmBvQa09O50U https://www.youtube.com/playlist?list=PL1D6Led19hjDG61mPEC8Wii-AKMHOCtfO https://www.youtube.com/playlist?list=PLBLO37Hy6RNAUMgYwQErjaDuI-EAlDfdE https://www.youtube.com/playlist?list=PLN5HOHzLUbahEWq8YAM8WhD-4Y58ANPp- https://www.youtube.com/playlist?list=PL90A0HJl0Q_VSkHt71-0_JYKPU4CDYCqL https://www.youtube.com/playlist?list=PLtuKjXLNTK35Hk1mJ0DnsBS9W2wkm5TXe https://www.youtube.com/playlist?list=PLYU7T1QT5d_P04Rc4csQNgOq4XZn2J3UQ https://www.youtube.com/playlist?list=PLZNC0fOGfVEi2ncf0aX9f-DbSDIkF-lYi https://www.youtube.com/playlist?list=PLvwjeYnP3Hn5hz9bbijf_DeHqWFJiZ-2K
https://vk.com/parimatch_vhod_registraciya
[url=https://vk.com/parimatch_zerkalo_rabohcee_2021]Париматч зеркало последнее рабочее 2021[/url]
KendraEmurf
Салют Поддерживаю И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=PQu5z8kzS90 https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8 https://www.youtube.com/shorts/UQR6a9gnt1s
https://www.youtube.com/playlist?list=PLRcnUiG5ZvBA76sZ5HKOBNu1WlZ9EWTMA https://www.youtube.com/playlist?list=PLRcnUiG5ZvBDF0EqqcL85ykxv9oRhTug1 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBzLEf09gytgCXRvjj8r0Sd https://www.youtube.com/playlist?list=PLRcnUiG5ZvBCgjuEcT4S7PeffoF-dLd9d https://www.youtube.com/playlist?list=PLRcnUiG5ZvBCCvjLL7rOgFt-Nt1WaDbq6 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBJSevb7fwp3zVa7hDjMZLd https://www.youtube.com/playlist?list=PLRcnUiG5ZvBD87ko1v1hrlsZHXMMWFcb2 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBD-Easch7fJFm7mbh30t8Qd
GedInsof
help for teens
http://historymed.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://tubesweet.xyz/categories http://ets-k.kz/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
Gennie
I blog quite often and I genuinely appreciate your information. This article has really peaked my interest.
I’m going to book mark your site and keep checking for new details about once a week. I opted in for your RSS feed too.
Reyna
I have been exploring for a little for any high-quality articles or weblog posts in this sort of space . Exploring in Yahoo I eventually stumbled upon this web site. Studying this info So i am satisfied to show that I’ve a very just right uncanny feeling I came upon just what I needed. I most indubitably will make sure to don?t put out of your mind this site and provides it a glance on a continuing basis.
Damian
Hello to all, the contents present at this web page are in fact awesome for people experience, well, keep up the nice work fellows.
Forrest
This site was… how do I say it? Relevant!! Finally I’ve found something that helped me. Kudos!
Elena
I am not certain where you are getting your information, however great topic. I must spend a while finding out more or figuring out more. Thank you for magnificent info I was searching for this information for my mission.
Matilda
Ahaa, its fastidious conversation regarding this piece of writing here at this website, I have read all that, so now me also commenting at this place.
JoanneHek
Привет согласен со всем И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/shorts/5Gmus2mjrVg https://www.youtube.com/watch?v=zXdEF-l1uS4 https://www.youtube.com/watch?v=apGB0wiCnEc https://www.youtube.com/watch?v=GXPDVU4uXg4 https://www.youtube.com/watch?v=eDpG1G9bGko https://www.youtube.com/watch?v=g_mHm8WGEl4 https://www.youtube.com/shorts/_bUDfPk9iUY https://www.youtube.com/watch?v=WMY5x8uds3Y https://www.youtube.com/watch?v=tqrBVSmvglM https://www.youtube.com/watch?v=wxyLpD-Qfu8
[url=https://www.youtube.com/watch?v=6_Tf-kz_Ljs]1XBET зеркало рабочий домен на сегодня сейчас 1ХБЕТ работающий адрес сайта скачать ios 2022[/url] [url=https://www.youtube.com/watch?v=6LYHvzd3xwM]Как скачать приложение 1xbet зеркало на андроид 4.4.2 бесплатно русском android 1хбет 2022[/url] [url=https://www.youtube.com/watch?v=vH7GkC1L3po]Как скачать приложение 1xbet зеркало на андроид 4.4.2 бесплатно русском android 1хбет[/url] [url=https://www.youtube.com/watch?v=Rr47JoUXnQM]Как скачать 1xbet на Андроид последняя версия зеркало работающее официальный сайт казино[/url] [url=https://www.youtube.com/watch?v=LnMDBErfRnA]1xbet зеркало сайта вход в личный кабинет рабочее на сегодня прямо сейчас 1хбет зеркало[/url] [url=https://www.youtube.com/watch?v=cgZRqrf84KM]1xbet зеркало сайта вход в личный кабинет рабочее на сегодня прямо сейчас 1хбет зеркало[/url] [url=https://www.youtube.com/shorts/5Gmus2mjrVg]1xbet зеркало рабочее на сегодня вход сайт 1хбет казино по смс срочно скачать через вк[/url]
[url=https://www.youtube.com/playlist?list=PLszZLCM3hGlHliHKiGnDB8bfzaY216lOE]1XBET bangla tutorial promo code - 1xbet Deposit Problem 2022 1xbet Deposit Rejected 2022 1xbet Deposit Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLvdaRBR-pJDc4RV2Yv–fn8dx48Crf5nt]1XBET promo code bangla - 1xBet promo code Bangladesh. Promo code 1xBet Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLfRfVVrJ6cc6T4OxWAkpi5tkuQmrwgkPD]Promo Code ?? 1xbet Promo Code use 1xbet Promo Code Bangla Video 1xbet Promo Code BD - 1xbet promo code bangladesh 2022[/url] [url=https://www.youtube.com/playlist?list=PLyffccC-f9_zwsN4gSD7xstT7kuIkILWS]1xbet Registration Bangla 2022 new app How To Create/Open 1xbet account - 1XBET Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLjQs__4gwB9WabwSvybkx_ZYGsnJdqhvm]1xBet promo code Bangladesh 1xbet Bangla Tutorial - 1XBET bangla tutorial promo code[/url] [url=https://www.youtube.com/playlist?list=PLGUAaPvNnfdyUHTu5xSiJuRujsL9tlf8F]Linebet bangla tutorial 2022 Linebet Better Than 1xbet Melbet - 1xbet promo code bangla android[/url] [url=https://www.youtube.com/playlist?list=PLGqzaqW711vIEI71zHSyW5JXA51Z7lck5]Best promo code for 1xbet Bangladesh and India 2022 free bet for registration Android - 1xbet promo code 2022 bangla[/url] [url=https://www.youtube.com/playlist?list=PLSbhbityRxhN8If0EW_oaa06fXcOuRJY7]1XBET promo code for registration - 1xbet Diposit problem 100% solve bkash, Nagad, bangladesh - best promo code for 1xbet bangladesh[/url] [url=https://www.youtube.com/playlist?list=PL1dppbSD4sxvVsYiC5k-YYHEVsLTjr9Sw]How to create 1xbet account bangla how to open 1xbet account 2022 1xbet registration bangla 1x - Promo code for 1XBET bangladesh[/url]
https://www.youtube.com/playlist?list=PL4PiX0z6gFYe7wnEWAc3hNrkfymCmJqhA https://www.youtube.com/playlist?list=PLHV7-qhrKACM0vuWoa5pYgtsT43KUO6ek https://www.youtube.com/playlist?list=PLhTL01CCZ4DrxcFQ5aJfpA1D3-VNsM1Kx https://www.youtube.com/playlist?list=PLPH-IC8r4kFEGrtyEkWRGqUnLrjAeHCUH https://www.youtube.com/playlist?list=PLnx2HACET3TOhJYcsw7TO0vL1JUuHagSB https://www.youtube.com/playlist?list=PLgwoHAwRWuSXfShVoou9gnczQ5RzhhZiI https://www.youtube.com/playlist?list=PLH5JHmNLTAzTE1yUFFqfF6mHUkWVee78D https://www.youtube.com/playlist?list=PLmsxJP-MOi-s-MRk2iwog9A_XWkJ2Vdfq https://www.youtube.com/playlist?list=PLAzjLuk66MHBgXWABVLB8WCoNA8Cf19ze
June
Hey there just wanted to give you a quick heads up. The words in your article seem to be running off the screen in Chrome. I’m not sure if this is a formatting issue or something to do with browser compatibility but I thought I’d post to let you know. The layout look great though! Hope you get the issue fixed soon. Kudos
JoanneHek
Привет со свем согласен и даже более Так же хочу добавит
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=PQu5z8kzS90 https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8 https://www.youtube.com/shorts/UQR6a9gnt1s
[url=https://www.youtube.com/watch?v=awnPK73IUEQ]Как скачать 1хбет зеркало на айфон - 1xbet на андроид рабочее на сегодня 2022[/url] [url=https://www.youtube.com/watch?v=cnFb5JF9I74]1xbet зеркало рабочее на сегодня прямо сейчас мобильная версия скачать айфон в онлайне 1хбет[/url [url=https://www.youtube.com/watch?v=Yo9WounjURA]Где взять 1xbet промокод при регистрации где смотреть если уже зарегистрирован 1хбет за регистрацию[/url] [url=https://www.youtube.com/watch?v=2VibXiXfzCo]Регистрация 1XBET промокод при регистрации 1хбет на бесплатную ставку 2022[/url] [url=https://www.youtube.com/watch?v=a4K_s3jd5sA]Как скачать 1хбет зеркало на айфон бесплатно 1xbet на андроид на русском зеркало #Shorts 2022[/url] [url=https://www.youtube.com/watch?v=HLrq-tjYTx8]Промокод 1XBET при регистрации на деньги 1хбет регистрация Промокод бесплатно Казахстан как получить[/url]
[url=https://www.youtube.com/playlist?list=PLF_zC7NDnQf-EW9AtuKx7RCZRzF2Bpwej]1xbet Bangladesh 2022 1xbet promo code Bangladesh 1xbet promo code Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLnfI0QSORsIiOhH7RT8V9zB3Ib_VR4vu7]1xbet promo code Bangla - 1x bet promo bonus code bangladesh 2022[/url] [url=https://www.youtube.com/playlist?list=PLKj6L6kYH-8pwaDYyglrqTMfBQtxZYDxK]1xbet Bangladesh 2022 1xbet promo code Bangladesh 1xbet promo code Bangladesh 2022[/url] [url=https://www.youtube.com/playlist?list=PL9KYoyRCRViCsI35vUwMCjVoLVhcrwMeo]1xbet promo code 2022 Bangla - 1xbet promo code promo code 1xbet 1xbet promo code 2022[/url] [url=https://www.youtube.com/playlist?list=PLwahMG6JXwgtu1I9klUmeNrj5aixt-EKa]1xbet how to open 1xbet account bangla.1xbet bangla tutorial.how to create 1xbet account.1xbet signup[/url] [url=https://www.youtube.com/playlist?list=PLmdhrMh2EqFKR10bddesFWgmEjmGRDvsl]How to create 1xbet account bangla how to open 1xbet account 2022 1xbet registration bangla 1x - Promo code for 1XBET bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLmX10wkAz_4bpPkr50-eNjWxY8OakalVH]1XBET promo code Bangladesh 2022 in Nepal Bangla code promo 1XBET 2022 English[/url] [url=https://www.youtube.com/playlist?list=PLmz65TI5qh4MGEtGC0qNVhpQ8PSYGMsKz]1XBET account registration bangladesh - Create a 1XBET Account Easily A-Z in Bangla Open 1xbet account 2022 Techzone Bangla[/url] [url=https://www.youtube.com/playlist?list=PLGwWlnPV-nr9IuU8OG4-Zj1WjGlvWUf6J]1XBET bangla tutorial promo code - 1xbet Deposit Problem 2022 1xbet Deposit Rejected 2022 1xbet Deposit Bangladesh[/url]
https://www.youtube.com/playlist?list=PL1JdbVJPQWNnk1Ttj_yL3Q38m45jc2frW https://www.youtube.com/playlist?list=PLkGIx6v36swDrrxLFXh9jOEx7KLuA6-Hw https://www.youtube.com/playlist?list=PLzRkediIjEJjQtvjX1v4fj7yR5A-wyK53 https://www.youtube.com/playlist?list=PLqy6OIpoxbZsNXnMvBvmc2sgIU3VUf8Sv https://www.youtube.com/playlist?list=PLiZmuOnZ385Vxzp4B43zvmahgNLLYvk4D https://www.youtube.com/playlist?list=PLQZHS9uvRNZjxpFxr8lbS1dXij0drUtil https://www.youtube.com/playlist?list=PLg_gU5dicCiyfs0FimRiYjtFtuSuVMtCN https://www.youtube.com/playlist?list=PLKQ2RMV8DaGtFxofO_7PKCWq8oI5ndNjY https://www.youtube.com/playlist?list=PLV6y5ivI1Fn-Dhd_qOKdCNCxZM7Wartgh
GedInsof
milf galleries
http://vsem-vizitki.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories http://fashion-fair.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://tubesweet.xyz/categories
JoanneHek
Салют со свем согласен Так вот
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=6_Tf-kz_Ljs https://www.youtube.com/watch?v=6LYHvzd3xwM https://www.youtube.com/watch?v=vH7GkC1L3po https://www.youtube.com/watch?v=Rr47JoUXnQM https://www.youtube.com/watch?v=LnMDBErfRnA https://www.youtube.com/watch?v=cgZRqrf84KM
[url=https://www.youtube.com/watch?v=LYnsyGU20qs]1xbet promo code biggest bonus in 2022 for registration account [/url] [url=https://www.youtube.com/watch?v=PQu5z8kzS90]1xbet promo code biggest bonus in 2022 for registration account [/url] [url=https://www.youtube.com/watch?v=rtX865F7jAo]1xbet me promo code kaise dale - comment creer son code promo 1xbet[/url] [url=https://www.youtube.com/watch?v=zi_rpdpjLo8]1xbet me promo code kaise dale - comment creer son code promo 1xbet [/url] [url=https://www.youtube.com/shorts/UQR6a9gnt1s]Как скачать 1хбет зеркало на айфон бесплатно 1xbet на андроид на русском зеркало 2022[/url]
[url=https://www.youtube.com/playlist?list=PLEBUODUctZO9eTz8LNHQLeYsTcXfpaA1B]Promo code 1xBet Bangladesh - 1XBET promo code bangla 1xBet promo code Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLT2jWTYv4X1auK8zsKdsa0xlxtV05zQt4]1x bet promo bonus code bangladesh 2022 - 1xbet promo code Bangla[/url] [url=https://www.youtube.com/playlist?list=PLrjJ6mR6zrZryGb8UM4uskhdl9qeZE414]1xbet promo code Bangladesh 2022 1xbet Bangladesh 2022 1xbet promo code Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLmjEqNxuviBSwC72aSIy4yOQzdFk0HekL]Promo Code ?? 1xbet Promo Code use 1xbet Promo Code Bangla Video 1xbet Promo Code BD - 1xbet promo code bangladesh 2022[/url] [url=https://www.youtube.com/playlist?list=PLooJc4ieNOfA2J8YTtRNEOQ3qh80TuK0I]How to Get Promo Points in 1XBET Bangla How to Play Promo Bet in 1XBET Melbet Free Promo Points - 1XBET promo code Bangladesh android[/url] [url=https://www.youtube.com/playlist?list=PLxBzrFl7ByegkbpykES7tjWLXyYAj2Ah6]1XBET promo code for registration - 1xbet Diposit problem 100% solve bkash, Nagad, bangladesh - best promo code for 1xbet bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLvF8VgIhx5ATFIDTi3ThGtV2diSPrHOKb]1xbet promo code hindi 1xbet promo code 1xbet promo code india, Pakistan, Bangladesh, nepal - 1xbet bangla tutorial promo code[/url] [url=https://www.youtube.com/playlist?list=PLGPmFPQotcMekAnyUUusNupoNAjk0Rt0T]1xbet Deposit Problem 2022 1xbet Deposit Rejected 2022 1xbet Deposit Bangladesh - 1XBET bangla tutorial promo code[/url]
https://www.youtube.com/playlist?list=PLqtP4xZb_S8-6688eHIQSXVh7skJ2MJ_F https://www.youtube.com/playlist?list=PLxxLGotVkhzImFcdqhtbnqVJ1xo6qctMB https://www.youtube.com/playlist?list=PL6hk1VT1OBHOt6QuObyQUkH0rcc6N4vNm https://www.youtube.com/playlist?list=PLNh5oggWgnBasrgxEiejFJQSooxddFsr1 https://www.youtube.com/playlist?list=PL3ePGZehRFcajul_U9wzQ-0GDQxxSTiuG https://www.youtube.com/playlist?list=PLxELrnQQgz1ah7vTzFR8iArZw176iM1QP https://www.youtube.com/playlist?list=PLu3hyhuw9GoHeJvWXVB4AkgNEGZILcHZp https://www.youtube.com/playlist?list=PLNh5oggWgnBasrgxEiejFJQSooxddFsr1
KendraEmurf
Здравствуйте хорошая мысль но Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
https://www.youtube.com/playlist?list=PLNzG2x9bf1wxkJXXWFQrI2Qq-8VHXgD1u https://www.youtube.com/playlist?list=PLNzG2x9bf1wwRwGZrF1GAY771VcANiV7y https://www.youtube.com/playlist?list=PLNzG2x9bf1wwP_ovG_-uF6hlQU-mQiZ2d https://www.youtube.com/playlist?list=PLNzG2x9bf1wy-k1qXGGu6URJzKrQC_QHN https://www.youtube.com/playlist?list=PLNzG2x9bf1wzdbxfnsIl9CKPPnK18JbXc https://www.youtube.com/playlist?list=PLNzG2x9bf1wxlt3We38spiaBkzJIxbnDI https://www.youtube.com/playlist?list=PLNzG2x9bf1wytmSbtsBC80Wyx1FrWF8d1 https://www.youtube.com/playlist?list=PLNzG2x9bf1wwHUZ0Ddknog6XlcoFTdN1j https://www.youtube.com/playlist?list=PLNzG2x9bf1wxLEtKqc8b0aEkdmmtcPAK8
PhillipITall
Hello there! [url=http://canadianxldrugstore.com/]online pharmacy vicodin[/url] great web page http://canadianxldrugstore.com
Williamden
pain meds online without doctor prescription prescription drugs without a doctor https://prescriptiondrugs24.com/# how can i order prescription drugs without a doctor
Clint
bookmarked!!, I really like your blog!
KendraEmurf
Hi согласен и даже более Так вот
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=cnFb5JF9I74 https://www.youtube.com/watch?v=Yo9WounjURA https://www.youtube.com/watch?v=2VibXiXfzCo https://www.youtube.com/watch?v=a4K_s3jd5sA https://www.youtube.com/watch?v=Ooe7Zhr8Ios https://www.youtube.com/watch?v=nAUbLZOMfU8 https://www.youtube.com/watch?v=MJhG3LuCWTw
https://www.youtube.com/playlist?list=PLneJ0QN_YNk6baz9zeudBhUCzYlGtPE_0 https://www.youtube.com/playlist?list=PLneJ0QN_YNk7Y8tx-VgSnl7ubzdIKCRnX https://www.youtube.com/playlist?list=PLneJ0QN_YNk5iRlC3MVh2RsAFmB88YSZE https://www.youtube.com/playlist?list=PLneJ0QN_YNk6OLDy8SaIvgWsMxEHQubOo https://www.youtube.com/playlist?list=PLneJ0QN_YNk7jFysr32Sf6Q5yu9B7eoH2 https://www.youtube.com/playlist?list=PLneJ0QN_YNk5ThXvfx3JOuKX-omxqpcHK https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBrBmHmAX_RHanGvmD464yf https://www.youtube.com/playlist?list=PLRcnUiG5ZvBAUZm9GihHfYfDK_Ftm7HoM
PhillipITall
Hello there! [url=http://canadianxldrugstore.com/]online us pharmacy[/url] very good web page http://canadianxldrugstore.com
KendraEmurf
Здравствуйте со свем согласен Так же по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
https://www.youtube.com/playlist?list=PLwdgj7Hr0nfcifMaaXdNLMdyjAqCn674m https://www.youtube.com/playlist?list=PLwdgj7Hr0nffUa0cUCRhRlJSMRYS1rRZU https://www.youtube.com/playlist?list=PLwdgj7Hr0nfe9s31ABJ20VRRiw4Tv2LjA https://www.youtube.com/playlist?list=PLwdgj7Hr0nfd5Q_k_o18OGohzAORueWtA https://www.youtube.com/playlist?list=PLwdgj7Hr0nff8VD0SzFvdtUx05gaIyOi4 https://www.youtube.com/playlist?list=PLwdgj7Hr0nfcuwzIjj-5ZR25IzJGXrYjU https://www.youtube.com/playlist?list=PLwdgj7Hr0nfeA-EX7hbjVCf7bfF7lg4V_ https://www.youtube.com/playlist?list=PLwdgj7Hr0nfdqijoADTcSiTBdL46p0Bcv
Horacio
Hi there it’s me, I am also visiting this website on a regular basis, this web page is in fact pleasant and the users are in fact sharing nice thoughts.
Yvonne
For newest news you have to pay a visit web and on web I found this web page as a best site for latest updates.
Corey
After going over a handful of the articles on your blog, I seriously like your way of blogging. I book-marked it to my bookmark website list and will be checking back soon. Please check out my web site too and let me know your opinion.
Kevinjailt
Hello there! [url=https://onlinepharmacyxls.com/]online pharmacies usa[/url] very good website https://onlinepharmacyxls.com
Alfie
you’re in point of fact a good webmaster. The website loading speed is incredible. It kind of feels that you are doing any distinctive trick. In addition, The contents are masterwork. you have performed a wonderful activity in this subject!
Marguerite
Thanks for sharing your info. I truly appreciate your efforts and I will be waiting for your further write ups thank you once again.
Eartha
you’re in reality a good webmaster. The website loading pace is amazing. It kind of feels that you’re doing any distinctive trick. Also, The contents are masterpiece. you’ve performed a fantastic job on this topic!
Victorina
Thanks for your marvelous posting! I definitely enjoyed reading it, you can be a great author.I will be sure to bookmark your blog and may come back very soon. I want to encourage continue your great job, have a nice holiday weekend!
Eddie
Very shortly this site will be famous among all blog viewers, due to it’s fastidious posts
Selina
I need to to thank you for this wonderful read!! I absolutely enjoyed every little bit of it.
I have you bookmarked to check out new stuff you post…
Joseph
I was curious if you ever thought of changing the layout of your website? Its very well written; I love what youve got to say. But maybe you could a little more in the way of content so people could connect with it better. Youve got an awful lot of text for only having one or two images.
Maybe you could space it out better?
Stanton
This design is spectacular! You obviously know how to keep a reader amused. Between your wit and your videos, I was almost moved to start my own blog (well, almost…HaHa!) Great job. I really enjoyed what you had to say, and more than that, how you presented it. Too cool!
Kevinjailt
Howdy! [url=https://onlinepharmacyxls.com/]pharmacy technician online courses[/url] very good website https://onlinepharmacyxls.com
JoanneHek
Салют со свем согласен и даже более Так же по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=cnFb5JF9I74 https://www.youtube.com/watch?v=Yo9WounjURA https://www.youtube.com/watch?v=2VibXiXfzCo https://www.youtube.com/watch?v=a4K_s3jd5sA https://www.youtube.com/watch?v=Ooe7Zhr8Ios https://www.youtube.com/watch?v=nAUbLZOMfU8 https://www.youtube.com/watch?v=MJhG3LuCWTw
[url=https://www.youtube.com/watch?v=Ooe7Zhr8Ios]1XBET регистрация по телефону и вход в личный кабинет как зарегистрироваться по номеру телефона[/url]
1xbet loss cover tricks 1xbet prediction Bangla Tutorial online tips Bangla[/url] [url=https://www.youtube.com/playlist?list=PLmsxJP-MOi-s-MRk2iwog9A_XWkJ2Vdfq]1xBet promo code Bangladesh 1xbet Bangla Tutorial - 1XBET bangla tutorial promo code[/url] [url=https://www.youtube.com/playlist?list=PLAzjLuk66MHBgXWABVLB8WCoNA8Cf19ze]1xbet.how to open 1xbet account bangla.1xbet bangla tutorial.how to create 1xbet account 1xbet signup[/url]
https://www.youtube.com/playlist?list=PL4Xvx92NaWgMq8rnRWCrbci-nwv1QYFZU https://www.youtube.com/playlist?list=PL7tNg1ikfbaf16962UBy3jEuSAZLglnp6 https://www.youtube.com/playlist?list=PLR08l22i117fUg3OgumlJmev7fz7NSs5k https://www.youtube.com/playlist?list=PLlpMriMa7LVTJ-QyxkjIFrdC0ResMfLb2 https://www.youtube.com/playlist?list=PL-j4gCqz4FaFyC7hAy6xHi0wPAL_t4AVK https://www.youtube.com/playlist?list=PLbBCkWf82N4YEDW-na2jwzN8IV2p3ZCOd https://www.youtube.com/playlist?list=PL_7gNaEvxtG-NkRhDn8kaxKBglaA4vOH6 https://www.youtube.com/playlist?list=PLY6rvNl-3tuhtYy-UOvTCo3dSsvhN20vl
Shelly
knewton coursework coursework uitm coursework in
KendraEmurf
Hi согласен и даже более Так вот
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=cnFb5JF9I74 https://www.youtube.com/watch?v=Yo9WounjURA https://www.youtube.com/watch?v=2VibXiXfzCo https://www.youtube.com/watch?v=a4K_s3jd5sA https://www.youtube.com/watch?v=Ooe7Zhr8Ios https://www.youtube.com/watch?v=nAUbLZOMfU8 https://www.youtube.com/watch?v=MJhG3LuCWTw
https://www.youtube.com/playlist?list=PLMC73QTysh692CUnCp2CXXUqaZ-9FDcKs https://www.youtube.com/playlist?list=PLMC73QTysh6-h_rN-Z8FPZgc4iN63tbiy https://www.youtube.com/playlist?list=PLMC73QTysh6-kjU0yKf2a0VGxzAcSfqlr https://www.youtube.com/playlist?list=PLMC73QTysh69gNeL0R83L29FylbyhRGUB https://www.youtube.com/playlist?list=PLMC73QTysh6-DEa-XrQtDIx4VIoL625pK https://www.youtube.com/playlist?list=PLMC73QTysh6_E6_0ygw9HBroZJ1p3cHyp https://www.youtube.com/playlist?list=PLMC73QTysh6-4-b7PUADZKqtfc126rrlo https://www.youtube.com/playlist?list=PLMC73QTysh68KqlL1Lfu6hsF-rBOpjz3q
Melina
coursework to order coursework sample coursework sample of written work
Randy
WOW just what I was searching for. Came here by searching for tft varus compo
Stuart
Asking questions are actually fastidious thing if you are not understanding something fully, however this post offers good understanding even.
Keesha
What’s up to every single one, it’s really a fastidious for me to pay a quick visit this web site, it includes important Information.
Kennith
The Zephyrus S17 has a big four cell, 90WHr battery. Four tweeters are positioned beneath the keyboard whereas the 2 woofers sit under the display. Overall, these two benchmark results bode well for avid gamers wanting a laptop computer that’s a reduce above by way of graphics performance, with the excessive frame rates equating to a smoother gaming experience and more detail in each scene rendered. Because the Xbox 360 cores can every handle two threads at a time, the 360 CPU is the equal of getting six standard processors in one machine. The sheer amount of calculating that is finished by the graphics processor to determine the angle and transparency for each mirrored object, and then render it in actual time, is extraordinary. Southern California artist Cosmo Wenman has used a 3-D printer to make meticulously rendered copies of well-known sculptures, based upon plans long-established from a whole bunch of photographs that he snaps from every angle. Many firms offer a combination of these plans. With its combination of a Core i9-11900H CPU, a Nvidia RTX 3080 GPU, and 32GB of RAM, this laptop carried out exceedingly effectively in performance benchmarks that provide perception into its CPU power and cooling. What this implies when you find yourself taking part in video video games is that the Xbox 360 can dedicate one core solely to producing sound, whereas another might run the sport’s collision and physics engine.
Oliver
It’s remarkable to pay a visit this website and reading the views of all colleagues on the topic of this piece of writing, while I am also keen of getting familiarity.
BradleyDex
Hi there! [url=http://finasteridexl.com/]buy finasteride[/url] very good website http://finasteridexl.com
Gavin
Thanks for sharing your thoughts about electronic cigarette vape. Regards
Lettie
Simply want to say your article is as surprising. The clearness in your post is just spectacular and i could assume you are an expert on this subject. Well with your permission let me to grab your feed to keep up to date with forthcoming post. Thanks a million and please keep up the enjoyable work.
Karen
Do you mind if I quote a couple of your articles as long as I provide credit and sources back to your website? My blog is in the exact same niche as yours and my users would truly benefit from some of the information you present here. Please let me know if this alright with you. Regards!
Jerry
Greetings! Very helpful advice in this particular article! It is the little changes which will make the greatest changes.
Thanks for sharing!
Emile
Hi to every one, the contents present at this web site are truly awesome for people knowledge, well, keep up the nice work fellows.
ThomasFam
cure ed: https://edpillsfast.com/
Charlie
I have read so many posts concerning the blogger lovers except this paragraph is actually a good piece of writing, keep it up.
Jeannie
My brother suggested I may like this blog. He was entirely right. This publish actually made my day. You can not consider simply how a lot time I had spent for this information! Thank you!
Cooper
I quite like reading an article that will make men and women think. Also, many thanks for permitting me to comment!
BradleyDex
Hi there! [url=http://finasteridexl.com/]generic propecia[/url] beneficial website http://finasteridexl.com
Princess
After going over a number of the articles on your blog, I seriously like your way of blogging. I saved it to my bookmark website list and will be checking back in the near future. Please check out my website as well and tell me what you think.
Senaida
coursework define coursework report coursework english language
Gladys
coursework resume example coursework help university coursework jamii forum
BradleyDex
Hi! [url=http://finasteridexl.com/]propecia price[/url] beneficial web page http://finasteridexl.com
Tanesha
Aw, this was a really good post. Finding the time and actual effort to create a really good article… but what can I say… I procrastinate a whole lot and never manage to get anything done.
Nathaniel
online coursework coursework and research difference coursework writing uk
Hannelore
Excellent website you have here but I was curious if you knew of any forums that cover the same topics talked about here? I’d really like to be a part of community where I can get opinions from other experienced people that share the same interest. If you have any recommendations, please let me know. Appreciate it!
BradleyDex
Hi there! [url=http://finasteridexl.com/]propecia[/url] good web site http://finasteridexl.com
Burton
I all the time used to study article in news papers but now as I am a user of web therefore from now I am using net for content, thanks to web.
Callum
ATM skimming is like identity theft for debit cards: Thieves use hidden electronics to steal the non-public info saved on your card and record your PIN quantity to entry all that arduous-earned money in your account. If ATM skimming is so critical and excessive-tech now, what dangers do we face with our debit and credit score playing cards in the future? Mobile bank card readers let clients make a digital swipe. And, as safety is at all times a problem in the case of sensitive credit card info, we’ll explore a few of the accusations that competitors have made against other merchandise. If the motherboard has onboard video, attempt to take away the video card fully and boot utilizing the onboard model.
Replacing the motherboard usually requires changing the heatsink and cooling fan, and will change the kind of RAM your pc wants, so you have got to do a little analysis to see what parts you’ll need to buy in this case.
Grant
My brother recommended I might like this web site.
He was totally right. This post actually made my day. You cann’t imagine simply how much time I had spent for this information! Thanks!
JoshuaLeach
Hello there! [url=http://propecia.site/]buy propecia medication[/url] excellent web page http://propecia.site
Alphonso
Hi, this weekend is pleasant in favor of me, for the reason that this occasion i am reading this wonderful educational article here at my home.
Jarred
Can I simply say what a comfort to uncover someone that actually understands what they are discussing over the internet.
You definitely know how to bring a problem to light and make it important.
More and more people need to read this and understand this side of your story. I was surprised that you are not more popular given that you surely have the gift.
Camille
Stephanie most just lately served as Global Head of CPS and can additionally be Co-Chair of Apollo Women Empower , Apollo’s women’s network.
JoshuaLeach
Hello there! [url=http://propecia.site/]propecia cheap[/url] beneficial internet site http://propecia.site
Katiefaf
Привет согласен со всем И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=kscXIwyW4go https://www.youtube.com/watch?v=q2jTlxSQops https://www.youtube.com/watch?v=hEeTU0TMaw8
[url=https://www.youtube.com/playlist?list=PLmN3RbVkfAMDmo9m1UL7VNCTSfkIPddNT]How to check 1xbet promo code 2022 - 2023 Nigeria Comment avoir le codepromo 1x bet generator[/url] [url=https://www.youtube.com/playlist?list=PLiJor_p8N4w60ZLHOTroGCX5IquvR2XzD]Davido 1xbet promo code 2022 - 2023 list Ghana for Nigeria in Nigeria free Spins[/url] [url=https://www.youtube.com/playlist?list=PL8vqYYAOOZICx2FRLMhc0GVPBbrGBC-OF]1xbet app promo code 2022 - 2023 India aujourd’hui for bet du jour register Nepal[/url] [url=https://www.youtube.com/playlist?list=PLTSY3tF6a1djiGitNwrWBANwuqrLHzfhW]Mark angel 1xbet promo code 2022 - 2023 terms and conditions sydney talker 1x bet pr omo code[/url] [url=https://www.youtube.com/playlist?list=PLK1nJH6HSPbiI6LQQi4khiyUATa8vBoPC]Macaroni 1xbet promo code Bangladesh 2022 - 2023 bonus free bet create Cameroon[/url] [url=https://www.youtube.com/playlist?list=PLPhIVhfLFvnC2CW7vt1iF8BME-XMx5wAV]Comment avoir son code promo 1xbet 2022 - 2023 how to use 1x bet free bet promocode bonus[/url] [url=https://www.youtube.com/playlist?list=PLmifGoUVAuZjNpwKctRgSasVjVi5L4kWC]How to get 1xbet promo code 2022 - 2023 georgia free spins register in cameroon[/url] [url=https://www.youtube.com/playlist?list=PLiHFMjprs5_QtdQ0zoIDe0uiR1PT9JBzq]Free 1xbet promo code 2022 - 2023 app no deposit for registration how can i get 1x bet promocode[/url] [url=https://www.youtube.com/playlist?list=PLdHqEDcpb2HuRZEGZ6XDgmWw785szrneJ]Comment creer un code promo 1xbet 2022 - 2023 registration India quelle est le codepromo de 1x bet[/url]
https://www.youtube.com/playlist?list=PLSVrj–ybQeUEsM6hF-JdAU_OABRThmDy https://www.youtube.com/playlist?list=PLojeCFKRqJyFKJ6njPle72OukCWnZclDZ https://www.youtube.com/playlist?list=PLywaCQJeA-Eb9kMvO0Q4UQ7nXZTI-_i1n https://www.youtube.com/playlist?list=PLTM-1R8j3i-2dTcdHcPibZS26LMVqBMTt https://www.youtube.com/playlist?list=PLtqGdTJkei8zfTxY6gvRUd03lPvXqKNQ- https://www.youtube.com/playlist?list=PLpiky9T0SbB79twvIqXx4joQYjnlU6NE8 https://www.youtube.com/playlist?list=PLow2PSDWI6VbRbVNpDPLLUeVJlW9ClIKL https://www.youtube.com/playlist?list=PLcDpQ-RbDRzrHVrzkmohdXmLViwiTsCDv https://www.youtube.com/playlist?list=PLOalfiVAiKTUtjFoBFhOibgNjsWVhRXAW
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url] [url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня [/url]
https://vk.com/zerkalo__vhod_alternativniy_sait https://vk.com/zerkalo_registraciya_1x https://vk.com/1x_zerkalo_rabotayuschee
Alina
coursework plagiarism checker do my coursework for me design technology coursework
Katiefaf
Hi Приятно найти единомыщленников Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=kscXIwyW4go https://www.youtube.com/watch?v=q2jTlxSQops https://www.youtube.com/watch?v=hEeTU0TMaw8
[url=https://www.youtube.com/playlist?list=PLnPz0vjesyHU37pUCBld_vWwv9ViifOyj]Exemple de code promo 1xbet 2022 - 2023 pour[/url] [url=https://www.youtube.com/playlist?list=PLcYWa4z3tFa0QWem6cGNcrE7UXy7Dn2JH]Meaning of code promo paris 1xbet 2022 - 2023 how to get[/url] [url=https://www.youtube.com/playlist?list=PL1XIGoSIvavrSZqpeSeuGle1VYtgeSnQn]Isbae u code promo 1xbet 2022 - 2023 canal plus how to activate 1x bet promocode quoi Nigeria[/url] [url=https://www.youtube.com/playlist?list=PLEtVeldfwRoXU-aGC_aUsBOVWLgffbQ4d]Oluwadolarz 1xbet promo code 2022 - 2023 how to get[/url] [url=https://www.youtube.com/playlist?list=PLeOa7r2Tsmbs9MIKhhOVlobhuDUKqWV5d]Broda shaggi 1xbet enter promo code 2022 - 2023 India bonus[/url] [url=https://www.youtube.com/playlist?list=PLbz-3qbsxgHutFC1VYN08dUx7L75OgCWQ]Babarex 1xbet promo code 2022 - 2023 de general davido 1 xbet[/url] [url=https://www.youtube.com/playlist?list=PL0R3jwcQqS1vpo1VW7IX87mLkWx8gCtm_]I need 1xbet promo code 2022 - 2023 near abuja 1xbet[/url] [url=https://www.youtube.com/playlist?list=PLDVFexeRlWHXk4tURcen_SeAeaAY0tugT]Mr funny 1xbet promo code 2022 - 2023 South Africa[/url]
https://www.youtube.com/playlist?list=PLdHqEDcpb2HuRZEGZ6XDgmWw785szrneJ https://www.youtube.com/playlist?list=PLljapY4SgkeGKkrgjLIrr0WUC3pq4PfET https://www.youtube.com/playlist?list=PL5nztcvO3yLgwARCDiUNWd932fwjGU1fI https://www.youtube.com/playlist?list=PLbz-3qbsxgHutFC1VYN08dUx7L75OgCWQ https://www.youtube.com/playlist?list=PLlTr0O1wHwxClIlBN5B3OCGn0H2ro_usx https://www.youtube.com/playlist?list=PLvslze0wAxBuWQTbVS0noKwfKc66WK46e https://www.youtube.com/playlist?list=PLynBo_9dKi0WLv3l11ZOoA812iLokS7Hf https://www.youtube.com/playlist?list=PLuKy3rrbnbeDVpR9kmO3j10eSfFxid2ys
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url] [url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня [/url]
https://vk.com/zerkalo_rabochee_1x https://vk.com/dostup_k_saitu
JoshuaLeach
Hello! [url=http://propecia.site/]propecia online[/url] good internet site http://propecia.site
Katiefaf
Салют со свем согласен Так же по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=kscXIwyW4go https://www.youtube.com/watch?v=q2jTlxSQops https://www.youtube.com/watch?v=hEeTU0TMaw8
[url=https://www.youtube.com/playlist?list=PLDVFexeRlWHXk4tURcen_SeAeaAY0tugT]Mr funny 1xbet promo code 2022 - 2023 South Africa[/url] [url=https://www.youtube.com/playlist?list=PLSVrj–ybQeUEsM6hF-JdAU_OABRThmDy]Macaroni 1xbet promo code Bangladesh 2022 - 2023 bonus[/url] [url=https://www.youtube.com/playlist?list=PLojeCFKRqJyFKJ6njPle72OukCWnZclDZ]Best 1xbet promo code 2022 - 2023 azerbaijan for taking part in the race free spins Casino[/url] [url=https://www.youtube.com/playlist?list=PLywaCQJeA-Eb9kMvO0Q4UQ7nXZTI-_i1n]Get 1xbet promo code 2022 - 2023 azerbaijan how to use 1x bet promocode Casino[/url] [url=https://www.youtube.com/playlist?list=PLTM-1R8j3i-2dTcdHcPibZS26LMVqBMTt]Code promo 1xbet 2022 - 2023 paris gratuit how to create 1x bet promocode queen officiel lasisi[/url] [url=https://www.youtube.com/playlist?list=PLtqGdTJkei8zfTxY6gvRUd03lPvXqKNQ-]Code promo 1xbet 2022 - 2023 pour l’inscription how to generate 1x bet official promocode Casino[/url] [url=https://www.youtube.com/playlist?list=PLpiky9T0SbB79twvIqXx4joQYjnlU6NE8]1xbet promo code 1xbet promo code for Registration 1xbet promo code In Hindi 2023 1xbet promo[/url] [url=https://www.youtube.com/playlist?list=PLow2PSDWI6VbRbVNpDPLLUeVJlW9ClIKL]Promo code 1xbet for registration. How to use 1XBET bonus code 1xBet promo code for registration[/url]
https://www.youtube.com/playlist?list=PLDVFexeRlWHXk4tURcen_SeAeaAY0tugT https://www.youtube.com/playlist?list=PLSVrj–ybQeUEsM6hF-JdAU_OABRThmDy https://www.youtube.com/playlist?list=PLojeCFKRqJyFKJ6njPle72OukCWnZclDZ https://www.youtube.com/playlist?list=PLywaCQJeA-Eb9kMvO0Q4UQ7nXZTI-_i1n https://www.youtube.com/playlist?list=PLTM-1R8j3i-2dTcdHcPibZS26LMVqBMTt https://www.youtube.com/playlist?list=PLtqGdTJkei8zfTxY6gvRUd03lPvXqKNQ- https://www.youtube.com/playlist?list=PLpiky9T0SbB79twvIqXx4joQYjnlU6NE8 https://www.youtube.com/playlist?list=PLow2PSDWI6VbRbVNpDPLLUeVJlW9ClIKL
[url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня[/url] [url=https://vk.com/bet_bonus_perviy_depozit]1хбет бонус на первый депозит 100% 1xbet условия[/url] [url=https://vk.com/zerkalo__vhod_alternativniy_sait]1xbet зеркало рабочее вход альтернативный сайт[/url]
https://vk.com/zerkalo__vhod_alternativniy_sait https://vk.com/zerkalo_registraciya_1x https://vk.com/1x_zerkalo_rabotayuschee
JoshuaLeach
Hello there! [url=http://propecia.site/]order propecia[/url] great web page http://propecia.site
TimothyLat
meds online without doctor prescription ed pills without doctor prescription https://images.google.bf/url?q=https://naijamoviez.com verified canadian pharmacies [url=https://images.google.mv/url?q=https://naijamoviez.com]top erectile dysfunction pills[/url] online medications
JoshuaLeach
Hi! [url=http://propecia.site/]order finasteride[/url] excellent site http://propecia.site
JoanneHek
Hi согласен и даже более Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=cnFb5JF9I74 https://www.youtube.com/watch?v=Yo9WounjURA https://www.youtube.com/watch?v=2VibXiXfzCo https://www.youtube.com/watch?v=a4K_s3jd5sA https://www.youtube.com/watch?v=Ooe7Zhr8Ios https://www.youtube.com/watch?v=nAUbLZOMfU8 https://www.youtube.com/watch?v=MJhG3LuCWTw
[url=https://www.youtube.com/watch?v=6_Tf-kz_Ljs]1XBET зеркало рабочий домен на сегодня сейчас 1ХБЕТ работающий адрес сайта скачать ios 2022[/url] [url=https://www.youtube.com/watch?v=6LYHvzd3xwM]Как скачать приложение 1xbet зеркало на андроид 4.4.2 бесплатно русском android 1хбет 2022[/url] [url=https://www.youtube.com/watch?v=vH7GkC1L3po]Как скачать приложение 1xbet зеркало на андроид 4.4.2 бесплатно русском android 1хбет[/url] [url=https://www.youtube.com/watch?v=Rr47JoUXnQM]Как скачать 1xbet на Андроид последняя версия зеркало работающее официальный сайт казино[/url] [url=https://www.youtube.com/watch?v=LnMDBErfRnA]1xbet зеркало сайта вход в личный кабинет рабочее на сегодня прямо сейчас 1хбет зеркало[/url] [url=https://www.youtube.com/watch?v=cgZRqrf84KM]1xbet зеркало сайта вход в личный кабинет рабочее на сегодня прямо сейчас 1хбет зеркало[/url] [url=https://www.youtube.com/shorts/5Gmus2mjrVg]1xbet зеркало рабочее на сегодня вход сайт 1хбет казино по смс срочно скачать через вк[/url]
[url=https://www.youtube.com/playlist?list=PL1JdbVJPQWNnk1Ttj_yL3Q38m45jc2frW]1xbet a to z bangla tutorial 2022.how to open 1xbet account bangla.how to create 1xbet account bd - How to open 1XBET account in Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLkGIx6v36swDrrxLFXh9jOEx7KLuA6-Hw]1xBet Friday Bonus Bangla 1xBet Friday Bonus Wigered Secret Tips How to Play 1xBet Bonus Bangla - 1xbet promo code for registration[/url] [url=https://www.youtube.com/playlist?list=PLzRkediIjEJjQtvjX1v4fj7yR5A-wyK53]1XBET promo code Bangladesh 2022 - How to withdraw 1xbet Bonus Bangla 2022 - ?????? ????? ???? ????? 1*et[/url] [url=https://www.youtube.com/playlist?list=PLqy6OIpoxbZsNXnMvBvmc2sgIU3VUf8Sv]1xbet prediction Bangla Tutorial 1xbet cricket No risk 1xbet loss cover tricks online tips Bangla - 1xbet telegram channel promo code Bangla[/url] [url=https://www.youtube.com/playlist?list=PLiZmuOnZ385Vxzp4B43zvmahgNLLYvk4D]Linebet bangla tutorial 2022 Linebet Better Than 1xbet Melbet - 1xbet promo code bangla android[/url] [url=https://www.youtube.com/playlist?list=PLQZHS9uvRNZjxpFxr8lbS1dXij0drUtil]How to become 1xbet melbet agent in Nepal, India , Bangaladesh , Pakistan etc - Best promo code for 1XBET Bangladesh[/url] [url=https://www.youtube.com/playlist?list=PLg_gU5dicCiyfs0FimRiYjtFtuSuVMtCN]How to online betting like - 1XBET promo code bangladesh 1xbet bangla tutorial 2022 Online Income Bangla Tutorial 2022[/url] [url=https://www.youtube.com/playlist?list=PLKQ2RMV8DaGtFxofO_7PKCWq8oI5ndNjY]1x - Promo code for 1XBET bangladesh How to create 1xbet account bangla how to open 1xbet account 2022 1xbet registration bangla[/url] [url=https://www.youtube.com/playlist?list=PLV6y5ivI1Fn-Dhd_qOKdCNCxZM7Wartgh]1xbet Registration Bangla 2022 new app How To Create/Open 1xbet account - 1XBET Bangladesh[/url]
https://www.youtube.com/playlist?list=PL4PiX0z6gFYe7wnEWAc3hNrkfymCmJqhA https://www.youtube.com/playlist?list=PLHV7-qhrKACM0vuWoa5pYgtsT43KUO6ek https://www.youtube.com/playlist?list=PLhTL01CCZ4DrxcFQ5aJfpA1D3-VNsM1Kx https://www.youtube.com/playlist?list=PLPH-IC8r4kFEGrtyEkWRGqUnLrjAeHCUH https://www.youtube.com/playlist?list=PLnx2HACET3TOhJYcsw7TO0vL1JUuHagSB https://www.youtube.com/playlist?list=PLgwoHAwRWuSXfShVoou9gnczQ5RzhhZiI https://www.youtube.com/playlist?list=PLH5JHmNLTAzTE1yUFFqfF6mHUkWVee78D https://www.youtube.com/playlist?list=PLmsxJP-MOi-s-MRk2iwog9A_XWkJ2Vdfq https://www.youtube.com/playlist?list=PLAzjLuk66MHBgXWABVLB8WCoNA8Cf19ze
Katiefaf
Hi со свем согласен и даже более И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=kscXIwyW4go https://www.youtube.com/watch?v=q2jTlxSQops https://www.youtube.com/watch?v=hEeTU0TMaw8
[url=https://www.youtube.com/playlist?list=PLmN3RbVkfAMDmo9m1UL7VNCTSfkIPddNT]How to check 1xbet promo code 2022 - 2023 Nigeria Comment avoir le codepromo 1x bet generator[/url] [url=https://www.youtube.com/playlist?list=PLiJor_p8N4w60ZLHOTroGCX5IquvR2XzD]Davido 1xbet promo code 2022 - 2023 list Ghana for Nigeria in Nigeria free Spins[/url] [url=https://www.youtube.com/playlist?list=PL8vqYYAOOZICx2FRLMhc0GVPBbrGBC-OF]1xbet app promo code 2022 - 2023 India aujourd’hui for bet du jour register Nepal[/url] [url=https://www.youtube.com/playlist?list=PLTSY3tF6a1djiGitNwrWBANwuqrLHzfhW]Mark angel 1xbet promo code 2022 - 2023 terms and conditions sydney talker 1x bet pr omo code[/url] [url=https://www.youtube.com/playlist?list=PLK1nJH6HSPbiI6LQQi4khiyUATa8vBoPC]Macaroni 1xbet promo code Bangladesh 2022 - 2023 bonus free bet create Cameroon[/url] [url=https://www.youtube.com/playlist?list=PLPhIVhfLFvnC2CW7vt1iF8BME-XMx5wAV]Comment avoir son code promo 1xbet 2022 - 2023 how to use 1x bet free bet promocode bonus[/url] [url=https://www.youtube.com/playlist?list=PLmifGoUVAuZjNpwKctRgSasVjVi5L4kWC]How to get 1xbet promo code 2022 - 2023 georgia free spins register in cameroon[/url] [url=https://www.youtube.com/playlist?list=PLiHFMjprs5_QtdQ0zoIDe0uiR1PT9JBzq]Free 1xbet promo code 2022 - 2023 app no deposit for registration how can i get 1x bet promocode[/url] [url=https://www.youtube.com/playlist?list=PLdHqEDcpb2HuRZEGZ6XDgmWw785szrneJ]Comment creer un code promo 1xbet 2022 - 2023 registration India quelle est le codepromo de 1x bet[/url]
https://www.youtube.com/playlist?list=PLDVFexeRlWHXk4tURcen_SeAeaAY0tugT https://www.youtube.com/playlist?list=PLSVrj–ybQeUEsM6hF-JdAU_OABRThmDy https://www.youtube.com/playlist?list=PLojeCFKRqJyFKJ6njPle72OukCWnZclDZ https://www.youtube.com/playlist?list=PLywaCQJeA-Eb9kMvO0Q4UQ7nXZTI-_i1n https://www.youtube.com/playlist?list=PLTM-1R8j3i-2dTcdHcPibZS26LMVqBMTt https://www.youtube.com/playlist?list=PLtqGdTJkei8zfTxY6gvRUd03lPvXqKNQ- https://www.youtube.com/playlist?list=PLpiky9T0SbB79twvIqXx4joQYjnlU6NE8 https://www.youtube.com/playlist?list=PLow2PSDWI6VbRbVNpDPLLUeVJlW9ClIKL
[url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня[/url] [url=https://vk.com/bet_bonus_perviy_depozit]1хбет бонус на первый депозит 100% 1xbet условия[/url] [url=https://vk.com/zerkalo__vhod_alternativniy_sait]1xbet зеркало рабочее вход альтернативный сайт[/url] https://vk.com/dostup_k_saitu https://vk.com/bet_bonus_perviy_depozit
volkovch
reading books is good http://book-sale.online
Katiefaf
Hi со свем согласен и даже более Так же хочу добавит
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=kscXIwyW4go https://www.youtube.com/watch?v=q2jTlxSQops https://www.youtube.com/watch?v=hEeTU0TMaw8
[url=https://www.youtube.com/playlist?list=PLGRcJOI2Vg33EQgKpZ4HvQE3_Ew7IBiGW]1xbet promo code for bonus Registration 1xBet[/url] [url=https://www.youtube.com/playlist?list=PLFs1-YUc95Zc2snc3i5RpZ-4P7LU5DpgQ]Promo code 1xbet for registration How to use 1XBET bonus code[/url] [url=https://www.youtube.com/playlist?list=PLXMaTklXSC5C9ApHmaRZSdeVO2XdRJb_2]1xbet Promo code 1xbet promo code for registration 1xbet promo code 2022 - 2023 How to use in HINDI[/url] [url=https://www.youtube.com/playlist?list=PL2Eq7ovYsL-Hpg9AemKuuOdABsQOmvkIP]1xbet promo code 1xbet promo code for Registration 1xbet promo code In Hindi 2023 1xbet promo[/url] [url=https://www.youtube.com/playlist?list=PLVfhDOhEFG-AXRdHNZ6JpvgkEUU1l1bsB]1xbet me registration kaise kare? 1xbet promo code 1xbet promo code for registration[/url] [url=https://www.youtube.com/playlist?list=PLfQOZGudwBY5BeyCBQVGk6M2PcWFvyZXY]How to use free official 1xbet bonus code - 1xbet promo code for registration in 2022[/url] [url=https://www.youtube.com/playlist?list=PL_QEn5AVEKRAyoY9G8AdCMbeAtKSfIcDB]1xBET registration How to create 1xbet account. 1xBET promo code 1xBet bonus code promo code for registration[/url] [url=https://www.youtube.com/playlist?list=PLVWV4z2tSh2Iq6ZtYhoiyTVJKCoa5GDx9]1xbet promo code for registration in 2022. How to use free official 1xbet bonus code[/url]
https://www.youtube.com/playlist?list=PLnPz0vjesyHU37pUCBld_vWwv9ViifOyj https://www.youtube.com/playlist?list=PLcYWa4z3tFa0QWem6cGNcrE7UXy7Dn2JH https://www.youtube.com/playlist?list=PL1XIGoSIvavrSZqpeSeuGle1VYtgeSnQn https://www.youtube.com/playlist?list=PLEtVeldfwRoXU-aGC_aUsBOVWLgffbQ4d https://www.youtube.com/playlist?list=PLeOa7r2Tsmbs9MIKhhOVlobhuDUKqWV5d https://www.youtube.com/playlist?list=PLbz-3qbsxgHutFC1VYN08dUx7L75OgCWQ https://www.youtube.com/playlist?list=PL0R3jwcQqS1vpo1VW7IX87mLkWx8gCtm_ https://www.youtube.com/playlist?list=PLDVFexeRlWHXk4tURcen_SeAeaAY0tugT
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url] [url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня [/url]
https://vk.com/zerkalo_rabochee_1x https://vk.com/dostup_k_saitu
JoshuaLeach
Hello there! [url=http://propecia.site/]buy propecia no prescription[/url] great internet site http://propecia.site
leopsferbab
https://krutiminst.ru - просмотры инстаграм накрутка
Raymundo
I think this is among the most vital information for me. And i’m happy studying your article. But should remark on some basic issues, The website taste is perfect, the articles is in reality nice : D. Good task, cheers
Jake
I all the time used to study paragraph in news papers but now as I am a user of net therefore from now I am using net for posts, thanks to web.
JoshuaLeach
Hi there! [url=http://propecia.site/]propecia canada[/url] very good internet site http://propecia.site
Kay
Hi! Someone in my Myspace group shared this website with us so I came to check it out. I’m definitely enjoying the information. I’m book-marking and will be tweeting this to my followers! Outstanding blog and wonderful design.
KendraEmurf
Здравствуйте согласен и даже более И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=PQu5z8kzS90 https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8 https://www.youtube.com/shorts/UQR6a9gnt1s
https://www.youtube.com/playlist?list=PLRcnUiG5ZvBA76sZ5HKOBNu1WlZ9EWTMA https://www.youtube.com/playlist?list=PLRcnUiG5ZvBDF0EqqcL85ykxv9oRhTug1 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBzLEf09gytgCXRvjj8r0Sd https://www.youtube.com/playlist?list=PLRcnUiG5ZvBCgjuEcT4S7PeffoF-dLd9d https://www.youtube.com/playlist?list=PLRcnUiG5ZvBCCvjLL7rOgFt-Nt1WaDbq6 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBJSevb7fwp3zVa7hDjMZLd https://www.youtube.com/playlist?list=PLRcnUiG5ZvBD87ko1v1hrlsZHXMMWFcb2 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBD-Easch7fJFm7mbh30t8Qd
Maple
After looking over a handful of the articles on your website, I truly appreciate your way of blogging. I book marked it to my bookmark webpage list and will be checking back soon. Take a look at my web site too and let me know how you feel.
KendraEmurf
Салют хорошая мысль но И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=6_Tf-kz_Ljs https://www.youtube.com/watch?v=6LYHvzd3xwM https://www.youtube.com/watch?v=vH7GkC1L3po https://www.youtube.com/watch?v=Rr47JoUXnQM https://www.youtube.com/watch?v=LnMDBErfRnA https://www.youtube.com/watch?v=cgZRqrf84KM
https://www.youtube.com/playlist?list=PLh5ePbJAzt9Z2Q-kd1cfH8whXDLa8AoBE https://www.youtube.com/playlist?list=PLh5ePbJAzt9Y1KknzKjGtLfI6lVSHiH1v https://www.youtube.com/playlist?list=PLh5ePbJAzt9a8jtUPckxW7JFa7XLFu3_q https://www.youtube.com/playlist?list=PLh5ePbJAzt9aG2SGl7bHoXcY3i3tS3CPj https://www.youtube.com/playlist?list=PLh5ePbJAzt9acqpGWMpsvoN4gyub2E0iM https://www.youtube.com/playlist?list=PLh5ePbJAzt9bOcr0VA2molbjJ7flvwE1n https://www.youtube.com/playlist?list=PLh5ePbJAzt9Y9NxwxfpQoIcY1gc738vGm https://www.youtube.com/playlist?list=PLh5ePbJAzt9aAarFWcDwSxx4_gO309qKD https://www.youtube.com/playlist?list=PLh5ePbJAzt9ZVVcXQmQxXKukwrO6qRao9
https://www.youtube.com/playlist?list=PLq4BI-2vvhBJFSm_zIWHnVxAyC7_grtyM https://www.youtube.com/playlist?list=PLq4BI-2vvhBLxpjDLTpoO1htzns24L7XQ https://www.youtube.com/playlist?list=PLq4BI-2vvhBIOkrg9Q5mQZ3HzN4QyieTq https://www.youtube.com/playlist?list=PLq4BI-2vvhBJORFMgQteH3OGrZFAQRruY https://www.youtube.com/playlist?list=PLq4BI-2vvhBJiG7pECb7QDPwpDPhJlfCh https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc
Octavio
It’s great that you are getting ideas from this paragraph as well as from our argument made at this time.
KendraEmurf
Здравствуйте согласен со всем Так же хочу добавит
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
https://www.youtube.com/playlist?list=PLwdgj7Hr0nfcifMaaXdNLMdyjAqCn674m https://www.youtube.com/playlist?list=PLwdgj7Hr0nffUa0cUCRhRlJSMRYS1rRZU https://www.youtube.com/playlist?list=PLwdgj7Hr0nfe9s31ABJ20VRRiw4Tv2LjA https://www.youtube.com/playlist?list=PLwdgj7Hr0nfd5Q_k_o18OGohzAORueWtA https://www.youtube.com/playlist?list=PLwdgj7Hr0nff8VD0SzFvdtUx05gaIyOi4 https://www.youtube.com/playlist?list=PLwdgj7Hr0nfcuwzIjj-5ZR25IzJGXrYjU https://www.youtube.com/playlist?list=PLwdgj7Hr0nfeA-EX7hbjVCf7bfF7lg4V_ https://www.youtube.com/playlist?list=PLwdgj7Hr0nfdqijoADTcSiTBdL46p0Bcv
https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJiG7pECb7QDPwpDPhJlfCh https://www.youtube.com/playlist?list=PLq4BI-2vvhBJORFMgQteH3OGrZFAQRruY https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBI-Ab6axauG7f0PMciZvhxo https://www.youtube.com/playlist?list=PLq4BI-2vvhBLxpjDLTpoO1htzns24L7XQ https://www.youtube.com/playlist?list=PLq4BI-2vvhBJfSYQYyuDb9Jy7Bdz155kS https://www.youtube.com/playlist?list=PLq4BI-2vvhBIwDotoD1zozfoxAT1vioi3
KendraEmurf
Здравствуйте хорошая мысль но И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
https://www.youtube.com/playlist?list=PLAgMBxPQhIJGJvZpGL5LgOWddUn5r2kMo https://www.youtube.com/playlist?list=PLAgMBxPQhIJFpa_N2qmyouzlSAxLVBlLt https://www.youtube.com/playlist?list=PLAgMBxPQhIJFlVL1RCs4eM1nafMM2wUHU https://www.youtube.com/playlist?list=PLAgMBxPQhIJGEsaqAGhcJLoRV4dR-9-mY https://www.youtube.com/playlist?list=PLAgMBxPQhIJG-5dEtL8ckFOTw1xCA5LMN https://www.youtube.com/playlist?list=PLAgMBxPQhIJEhD_1aa8-0G09f-Kohzvyf https://www.youtube.com/playlist?list=PLAgMBxPQhIJHiaF23p95K_M_Jo0t7u0SH https://www.youtube.com/playlist?list=PLAgMBxPQhIJG39IzPh4My_KvmiUaXS1DD https://www.youtube.com/playlist?list=PLAgMBxPQhIJHrMfwCc-XYplvXROAEU6dY
https://www.youtube.com/playlist?list=PLq4BI-2vvhBIqLGPBPTc5ajwQLKPt3qku https://www.youtube.com/playlist?list=PLq4BI-2vvhBKPwrIEUEFeHH3KoXotByio https://www.youtube.com/playlist?list=PLq4BI-2vvhBIDVcvQLRwU3cr_gheV1qnp https://www.youtube.com/playlist?list=PLq4BI-2vvhBLoRRNWdgFMRGjxSdNaCWsC https://www.youtube.com/playlist?list=PLq4BI-2vvhBLoYurr-FLynMsXOGf7fCss https://www.youtube.com/playlist?list=PLq4BI-2vvhBKwAyyBQVZt4mRfpd2MBzF9 https://www.youtube.com/playlist?list=PLq4BI-2vvhBLrp8mENpEIsurTdBe1JxGE https://www.youtube.com/playlist?list=PLq4BI-2vvhBI3uain9fUkozCGiJUu_u3g https://www.youtube.com/playlist?list=PLq4BI-2vvhBIPuVjSapf4tMbvMls8Rpfr
JoshuaLeach
Hi there! [url=http://propecia.site/]buy propecia pills[/url] good website http://propecia.site
Katiefaf
Hi хорошая мысль но Так же по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=kscXIwyW4go https://www.youtube.com/watch?v=q2jTlxSQops https://www.youtube.com/watch?v=hEeTU0TMaw8
[url=https://www.youtube.com/playlist?list=PLGRcJOI2Vg33EQgKpZ4HvQE3_Ew7IBiGW]1xbet promo code for bonus Registration 1xBet[/url] [url=https://www.youtube.com/playlist?list=PLFs1-YUc95Zc2snc3i5RpZ-4P7LU5DpgQ]Promo code 1xbet for registration How to use 1XBET bonus code[/url] [url=https://www.youtube.com/playlist?list=PLXMaTklXSC5C9ApHmaRZSdeVO2XdRJb_2]1xbet Promo code 1xbet promo code for registration 1xbet promo code 2022 - 2023 How to use in HINDI[/url] [url=https://www.youtube.com/playlist?list=PL2Eq7ovYsL-Hpg9AemKuuOdABsQOmvkIP]1xbet promo code 1xbet promo code for Registration 1xbet promo code In Hindi 2023 1xbet promo[/url] [url=https://www.youtube.com/playlist?list=PLVfhDOhEFG-AXRdHNZ6JpvgkEUU1l1bsB]1xbet me registration kaise kare? 1xbet promo code 1xbet promo code for registration[/url] [url=https://www.youtube.com/playlist?list=PLfQOZGudwBY5BeyCBQVGk6M2PcWFvyZXY]How to use free official 1xbet bonus code - 1xbet promo code for registration in 2022[/url] [url=https://www.youtube.com/playlist?list=PL_QEn5AVEKRAyoY9G8AdCMbeAtKSfIcDB]1xBET registration How to create 1xbet account. 1xBET promo code 1xBet bonus code promo code for registration[/url] [url=https://www.youtube.com/playlist?list=PLVWV4z2tSh2Iq6ZtYhoiyTVJKCoa5GDx9]1xbet promo code for registration in 2022. How to use free official 1xbet bonus code[/url]
https://www.youtube.com/playlist?list=PLC13uStEaZ4DwOlvYJtK-ot5Wojhf9vty https://www.youtube.com/playlist?list=PLmN3RbVkfAMDmo9m1UL7VNCTSfkIPddNT https://www.youtube.com/playlist?list=PLiJor_p8N4w60ZLHOTroGCX5IquvR2XzD https://www.youtube.com/playlist?list=PL8vqYYAOOZICx2FRLMhc0GVPBbrGBC-OF https://www.youtube.com/playlist?list=PLTSY3tF6a1djiGitNwrWBANwuqrLHzfhW https://www.youtube.com/playlist?list=PLK1nJH6HSPbiI6LQQi4khiyUATa8vBoPC https://www.youtube.com/playlist?list=PLPhIVhfLFvnC2CW7vt1iF8BME-XMx5wAV https://www.youtube.com/playlist?list=PLmifGoUVAuZjNpwKctRgSasVjVi5L4kWC https://www.youtube.com/playlist?list=PLiHFMjprs5_QtdQ0zoIDe0uiR1PT9JBzq
[url=https://vk.com/zerkalo_registraciya_1x]ЗЕРКАЛО 1xbet регистрация на сайте в один клик[/url] [url=https://vk.com/1x_zerkalo_rabotayuschee]Зеркало 1xbet работающее на сегодня прямо сейчас[/url]
https://vk.com/zerkalo_rabochee_1x https://vk.com/dostup_k_saitu
Katiefaf
Салют со свем согласен Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=kscXIwyW4go https://www.youtube.com/watch?v=q2jTlxSQops https://www.youtube.com/watch?v=hEeTU0TMaw8
[url=https://www.youtube.com/playlist?list=PLDVFexeRlWHXk4tURcen_SeAeaAY0tugT]Mr funny 1xbet promo code 2022 - 2023 South Africa[/url] [url=https://www.youtube.com/playlist?list=PLSVrj–ybQeUEsM6hF-JdAU_OABRThmDy]Macaroni 1xbet promo code Bangladesh 2022 - 2023 bonus[/url] [url=https://www.youtube.com/playlist?list=PLojeCFKRqJyFKJ6njPle72OukCWnZclDZ]Best 1xbet promo code 2022 - 2023 azerbaijan for taking part in the race free spins Casino[/url] [url=https://www.youtube.com/playlist?list=PLywaCQJeA-Eb9kMvO0Q4UQ7nXZTI-_i1n]Get 1xbet promo code 2022 - 2023 azerbaijan how to use 1x bet promocode Casino[/url] [url=https://www.youtube.com/playlist?list=PLTM-1R8j3i-2dTcdHcPibZS26LMVqBMTt]Code promo 1xbet 2022 - 2023 paris gratuit how to create 1x bet promocode queen officiel lasisi[/url] [url=https://www.youtube.com/playlist?list=PLtqGdTJkei8zfTxY6gvRUd03lPvXqKNQ-]Code promo 1xbet 2022 - 2023 pour l’inscription how to generate 1x bet official promocode Casino[/url] [url=https://www.youtube.com/playlist?list=PLpiky9T0SbB79twvIqXx4joQYjnlU6NE8]1xbet promo code 1xbet promo code for Registration 1xbet promo code In Hindi 2023 1xbet promo[/url] [url=https://www.youtube.com/playlist?list=PLow2PSDWI6VbRbVNpDPLLUeVJlW9ClIKL]Promo code 1xbet for registration. How to use 1XBET bonus code 1xBet promo code for registration[/url] https://www.youtube.com/playlist?list=PLmN3RbVkfAMDmo9m1UL7VNCTSfkIPddNT https://www.youtube.com/playlist?list=PLiJor_p8N4w60ZLHOTroGCX5IquvR2XzD https://www.youtube.com/playlist?list=PL8vqYYAOOZICx2FRLMhc0GVPBbrGBC-OF https://www.youtube.com/playlist?list=PLTSY3tF6a1djiGitNwrWBANwuqrLHzfhW https://www.youtube.com/playlist?list=PLK1nJH6HSPbiI6LQQi4khiyUATa8vBoPC https://www.youtube.com/playlist?list=PLPhIVhfLFvnC2CW7vt1iF8BME-XMx5wAV https://www.youtube.com/playlist?list=PLmifGoUVAuZjNpwKctRgSasVjVi5L4kWC https://www.youtube.com/playlist?list=PLiHFMjprs5_QtdQ0zoIDe0uiR1PT9JBzq https://www.youtube.com/playlist?list=PLdHqEDcpb2HuRZEGZ6XDgmWw785szrneJ
[url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня[/url] [url=https://vk.com/bet_bonus_perviy_depozit]1хбет бонус на первый депозит 100% 1xbet условия[/url] [url=https://vk.com/zerkalo__vhod_alternativniy_sait]1xbet зеркало рабочее вход альтернативный сайт[/url] https://vk.com/dostup_k_saitu https://vk.com/bet_bonus_perviy_depozit
JoshuaLeach
Hello! [url=http://propecia.site/]finasteride 5mg[/url] good web site http://propecia.site
Shiela
It’s a shame you don’t have a donate button! I’d certainly donate to this excellent blog! I guess for now i’ll settle for book-marking and adding your RSS feed to my Google account.
I look forward to brand new updates and will share this website with my Facebook group. Talk soon!
Sanora
My brother suggested I might like this blog. He was entirely right. This post truly made my day. You can not imagine just how much time I had spent for this information! Thanks!
Meghan
You actually make it seem so easy with your presentation but I find this matter to be actually something which I think I would never understand. It seems too complicated and extremely broad for me. I am looking forward for your next post, I will try to get the hang of it!
KendraEmurf
Hi хорошая мысль но И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
https://www.youtube.com/playlist?list=PLneJ0QN_YNk5ULpCgAhMN9U3v81wiIn8S https://www.youtube.com/playlist?list=PLneJ0QN_YNk41flytvb9sWU5JTs6hcxfP https://www.youtube.com/playlist?list=PLneJ0QN_YNk4Zusz1_1dgbXeXgseMEOSO https://www.youtube.com/playlist?list=PLneJ0QN_YNk6rxCx20U_NywIFLeEfFHPx https://www.youtube.com/playlist?list=PLneJ0QN_YNk6gyggk4NIwX_5_bBgt7gLG https://www.youtube.com/playlist?list=PLneJ0QN_YNk4ZSS9rz5-KjdPEMCh1gK-W https://www.youtube.com/playlist?list=PLneJ0QN_YNk60vQU3WjmR51gB2h3o7XCw https://www.youtube.com/playlist?list=PLneJ0QN_YNk5vQGg3wjCLGYZZYjkcnKOB https://www.youtube.com/playlist?list=PLneJ0QN_YNk6kF0CYJX1TkYF7bVbWY6hl
https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJiG7pECb7QDPwpDPhJlfCh https://www.youtube.com/playlist?list=PLq4BI-2vvhBJORFMgQteH3OGrZFAQRruY https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBI-Ab6axauG7f0PMciZvhxo https://www.youtube.com/playlist?list=PLq4BI-2vvhBLxpjDLTpoO1htzns24L7XQ https://www.youtube.com/playlist?list=PLq4BI-2vvhBJfSYQYyuDb9Jy7Bdz155kS https://www.youtube.com/playlist?list=PLq4BI-2vvhBIwDotoD1zozfoxAT1vioi3
KendraEmurf
Здрасте согласен и даже более Так же по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=6_Tf-kz_Ljs https://www.youtube.com/watch?v=6LYHvzd3xwM https://www.youtube.com/watch?v=vH7GkC1L3po https://www.youtube.com/watch?v=Rr47JoUXnQM https://www.youtube.com/watch?v=LnMDBErfRnA https://www.youtube.com/watch?v=cgZRqrf84KM
https://www.youtube.com/playlist?list=PLAgMBxPQhIJGJvZpGL5LgOWddUn5r2kMo https://www.youtube.com/playlist?list=PLAgMBxPQhIJFpa_N2qmyouzlSAxLVBlLt https://www.youtube.com/playlist?list=PLAgMBxPQhIJFlVL1RCs4eM1nafMM2wUHU https://www.youtube.com/playlist?list=PLAgMBxPQhIJGEsaqAGhcJLoRV4dR-9-mY https://www.youtube.com/playlist?list=PLAgMBxPQhIJG-5dEtL8ckFOTw1xCA5LMN https://www.youtube.com/playlist?list=PLAgMBxPQhIJEhD_1aa8-0G09f-Kohzvyf https://www.youtube.com/playlist?list=PLAgMBxPQhIJHiaF23p95K_M_Jo0t7u0SH https://www.youtube.com/playlist?list=PLAgMBxPQhIJG39IzPh4My_KvmiUaXS1DD https://www.youtube.com/playlist?list=PLAgMBxPQhIJHrMfwCc-XYplvXROAEU6dY
https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJiG7pECb7QDPwpDPhJlfCh https://www.youtube.com/playlist?list=PLq4BI-2vvhBJORFMgQteH3OGrZFAQRruY https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBI-Ab6axauG7f0PMciZvhxo https://www.youtube.com/playlist?list=PLq4BI-2vvhBLxpjDLTpoO1htzns24L7XQ https://www.youtube.com/playlist?list=PLq4BI-2vvhBJfSYQYyuDb9Jy7Bdz155kS https://www.youtube.com/playlist?list=PLq4BI-2vvhBIwDotoD1zozfoxAT1vioi3
KendraEmurf
Здравствуйте со свем согласен И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=ZGiZvXquPcQ https://www.youtube.com/watch?v=g_d0Zyl2VqE https://www.youtube.com/watch?v=vIZD9oOl0tg https://www.youtube.com/watch?v=HLrq-tjYTx8 https://www.youtube.com/watch?v=TMBWt3Xf5xw https://www.youtube.com/watch?v=fieWXlMi01o https://www.youtube.com/watch?v=GhBNvgzg5oc
https://www.youtube.com/playlist?list=PLNzG2x9bf1wxkJXXWFQrI2Qq-8VHXgD1u https://www.youtube.com/playlist?list=PLNzG2x9bf1wwRwGZrF1GAY771VcANiV7y https://www.youtube.com/playlist?list=PLNzG2x9bf1wwP_ovG_-uF6hlQU-mQiZ2d https://www.youtube.com/playlist?list=PLNzG2x9bf1wy-k1qXGGu6URJzKrQC_QHN https://www.youtube.com/playlist?list=PLNzG2x9bf1wzdbxfnsIl9CKPPnK18JbXc https://www.youtube.com/playlist?list=PLNzG2x9bf1wxlt3We38spiaBkzJIxbnDI https://www.youtube.com/playlist?list=PLNzG2x9bf1wytmSbtsBC80Wyx1FrWF8d1 https://www.youtube.com/playlist?list=PLNzG2x9bf1wwHUZ0Ddknog6XlcoFTdN1j https://www.youtube.com/playlist?list=PLNzG2x9bf1wxLEtKqc8b0aEkdmmtcPAK8
https://www.youtube.com/playlist?list=PLq4BI-2vvhBKpthxPvURqkFnAAusp5gxu https://www.youtube.com/playlist?list=PLq4BI-2vvhBLuFrOMIPPw8ulahnfk6X94 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJng2iNLNFPQ0l6HLq5keQD https://www.youtube.com/playlist?list=PLq4BI-2vvhBKIH5Cy18DUUAuePgHYeNXX https://www.youtube.com/playlist?list=PLq4BI-2vvhBKnv_CwIoCcDypasoI5d4Tc https://www.youtube.com/playlist?list=PLq4BI-2vvhBLMogknP6oCK5QwUwo4AfWM
KendraEmurf
Hi Приятно найти единомыщленников Так же хочу добавит
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
https://www.youtube.com/playlist?list=PLRcnUiG5ZvBA76sZ5HKOBNu1WlZ9EWTMA https://www.youtube.com/playlist?list=PLRcnUiG5ZvBDF0EqqcL85ykxv9oRhTug1 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBzLEf09gytgCXRvjj8r0Sd https://www.youtube.com/playlist?list=PLRcnUiG5ZvBCgjuEcT4S7PeffoF-dLd9d https://www.youtube.com/playlist?list=PLRcnUiG5ZvBCCvjLL7rOgFt-Nt1WaDbq6 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBJSevb7fwp3zVa7hDjMZLd https://www.youtube.com/playlist?list=PLRcnUiG5ZvBD87ko1v1hrlsZHXMMWFcb2 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBD-Easch7fJFm7mbh30t8Qd
https://www.youtube.com/playlist?list=PLq4BI-2vvhBIqLGPBPTc5ajwQLKPt3qku https://www.youtube.com/playlist?list=PLq4BI-2vvhBKPwrIEUEFeHH3KoXotByio https://www.youtube.com/playlist?list=PLq4BI-2vvhBIDVcvQLRwU3cr_gheV1qnp https://www.youtube.com/playlist?list=PLq4BI-2vvhBLoRRNWdgFMRGjxSdNaCWsC https://www.youtube.com/playlist?list=PLq4BI-2vvhBLoYurr-FLynMsXOGf7fCss https://www.youtube.com/playlist?list=PLq4BI-2vvhBKwAyyBQVZt4mRfpd2MBzF9 https://www.youtube.com/playlist?list=PLq4BI-2vvhBLrp8mENpEIsurTdBe1JxGE https://www.youtube.com/playlist?list=PLq4BI-2vvhBI3uain9fUkozCGiJUu_u3g https://www.youtube.com/playlist?list=PLq4BI-2vvhBIPuVjSapf4tMbvMls8Rpfr
KendraEmurf
Привет Приятно найти единомыщленников Так же хочу добавит
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
https://www.youtube.com/playlist?list=PLNzG2x9bf1wwlbMby1ccV9htGRMfimWQH https://www.youtube.com/playlist?list=PLNzG2x9bf1wyjCzuyPrDuDytj9WQCYorr https://www.youtube.com/playlist?list=PLneJ0QN_YNk7h8FCoR5PDaQIWe7fmrE-z https://www.youtube.com/playlist?list=PLneJ0QN_YNk7iaBy95I0GkhDaZzRjqrjc https://www.youtube.com/playlist?list=PLneJ0QN_YNk4Nlh2_RqfdJ5PTNdjQ1Go- https://www.youtube.com/playlist?list=PLneJ0QN_YNk7BkdRH73blRJAzAekfneuk https://www.youtube.com/playlist?list=PLneJ0QN_YNk4E8VmkBR9Mix92IoXC7WDK https://www.youtube.com/playlist?list=PLneJ0QN_YNk5VsHxzDYTkDhtoC0DZf2uv
https://www.youtube.com/playlist?list=PLq4BI-2vvhBKpthxPvURqkFnAAusp5gxu https://www.youtube.com/playlist?list=PLq4BI-2vvhBLuFrOMIPPw8ulahnfk6X94 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJng2iNLNFPQ0l6HLq5keQD https://www.youtube.com/playlist?list=PLq4BI-2vvhBKIH5Cy18DUUAuePgHYeNXX https://www.youtube.com/playlist?list=PLq4BI-2vvhBKnv_CwIoCcDypasoI5d4Tc https://www.youtube.com/playlist?list=PLq4BI-2vvhBLMogknP6oCK5QwUwo4AfWM
KevinMix
Hello there! [url=http://uffcialis.top/]anyone buy cialis online[/url] very good web site http://uffcialis.online
Mckenzie
Incredible points. Great arguments. Keep up the good work.
WideRange
The at worst small EV charger with built-in ground in the extreme detection. We lone propose EV plug types based on recommended electrical code. No more disturbing breakers tripping. Our EV charger power settings do not pass the blurb’s capacity.
use this link http://ev-chargers.com/toyota/rav4/
[url=http://ev-chargers.com/nema-14-30/]your input here[/url]
<a href=https://ev-chargers.com/nema-14-30/>find</a>
Christie
Thanks for sharing your thoughts about electronic cigarettes for sale. Regards
Fiona
Hey there! Would you mind if I share your blog with my myspace group? There’s a lot of people that I think would really appreciate your content. Please let me know. Many thanks
Fabian
It’s a shame you don’t have a donate button! I’d certainly donate to this outstanding blog! I suppose for now i’ll settle for book-marking and adding your RSS feed to my Google account. I look forward to fresh updates and will talk about this blog with my Facebook group. Talk soon!
Yolanda
I’m not sure where you are getting your information, but good topic. I needs to spend some time learning much more or understanding more.
Thanks for wonderful info I was looking for this info for my mission.
Francine
A person essentially help to make critically posts I’d state. This is the first time I frequented your web page and thus far? I amazed with the research you made to make this actual put up amazing. Excellent process!
Valerie
Very good blog post. I certainly love this website. Continue the good work!
Lawanna
Appreciating the persistence you put into your blog and in depth information you present. It’s nice to come across a blog every once in a while that isn’t the same out of date rehashed information. Wonderful read! I’ve saved your site and I’m including your RSS feeds to my Google account.
Raphael
I’m curious to find out what blog system you have been using? I’m having some minor security issues with my latest blog and I’d like to find something more secure. Do you have any solutions?
KevinMix
Howdy! [url=http://uffcialis.top/]is it illegal to buy generic cialis online[/url] beneficial site http://uffcialis.online
Leatha
Hi, i think that i saw you visited my blog so i came to return the prefer?.I am attempting to find issues to improve my website!I assume its ok to make use of a few of your ideas!!
Reginaemeni
Здрасте Приятно найти единомыщленников Так вот
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=6_Tf-kz_Ljs https://www.youtube.com/watch?v=6LYHvzd3xwM https://www.youtube.com/watch?v=vH7GkC1L3po https://www.youtube.com/watch?v=Rr47JoUXnQM https://www.youtube.com/watch?v=LnMDBErfRnA https://www.youtube.com/watch?v=cgZRqrf84KM
[url=https://www.youtube.com/watch?v=nAUbLZOMfU8]1XBET регистрация по телефону и вход в личный кабинет как зарегистрироваться по номеру телефона[/url] [url=https://www.youtube.com/watch?v=MJhG3LuCWTw]1xbet регистрация Казахстан вход в личный кабинет Казакша вход в личный кабинет в 1хбет - скачать[/url] [url=https://www.youtube.com/watch?v=ZGiZvXquPcQ]1xbet регистрация килиш по номеру телефона для компьютера личный кабинет[/url] [url=https://www.youtube.com/watch?v=g_d0Zyl2VqE]1XBET зеркало регистрация в 1 клик по номеру телефона в один клик вход 1ХБЕТ[/url] [url=https://www.youtube.com/watch?v=vIZD9oOl0tg]1XBET зеркало регистрация в 1 клик по номеру телефона в один клик вход 1ХБЕТ[/url] [url=https://www.youtube.com/watch?v=TMBWt3Xf5xw]1xbet регистрация промокод при регистрации 1хбет 2022 [/url] [url=https://www.youtube.com/watch?v=fieWXlMi01o]1xbet ?алай регистрация жасайды іспит иностранных граждан в России[/url] [url=https://www.youtube.com/watch?v=GhBNvgzg5oc]Как узнать свой промокод 1xbet как добавить промо код в 1хбет на 2022 бк[/url]
https://www.youtube.com/playlist?list=PLlYfusLR2lbG-eL7V-haJG1rErgTENUA7 https://www.youtube.com/playlist?list=PLyoNgQjsK1zCP-tCPD8J5efouOx0PLlpR https://www.youtube.com/playlist?list=PLr1R4nt3KsmyXFgiCR3I_XJL5_1xM5gTE https://www.youtube.com/playlist?list=PLOqCIUKw9lSuJ8ayVEjv125u72qE6GoVt https://www.youtube.com/playlist?list=PLQhpdzZwqn3B9aU4AKQr4R5GYZm0eHMj5 https://www.youtube.com/playlist?list=PLBchsNgwfysWO8Xp-ixEHovv02Z7fW43z https://www.youtube.com/playlist?list=PL6tTAH7k1elefRCVdDyooCfNcbf4CS6eh https://www.youtube.com/playlist?list=PLe8ZPPuaNGnmCXNNCFMW8HKykm4N0o5e1 https://www.youtube.com/playlist?list=PL8h99E2aNCjDLsa0VbIh30Hf_OnbD-6dB
[url=https://www.youtube.com/playlist?list=PLNoAkrN9jep79VONfM37oX_aO06vYBFsz]1xbet актуальное рабочее зеркало альтернативный адрес на сегодня на май 2022[/url] [url=https://www.youtube.com/playlist?list=PLVXbMH_gmerWir4S2a4Ix4iVejt9sRZL_]1xbet рабочее зеркало на сегодня прямо сейчас 1хбет 2022[/url] [url=https://www.youtube.com/playlist?list=PLPPJ1hx14qAhlmppprSGnjJhe3krNF9SK]1xbet рабочее зеркало на сегодня прямо сейчас 1хбет скачать на андроид бесплатно на русском в мае месчце 2022 год[/url] [url=https://www.youtube.com/playlist?list=PLQI1Vc8W_P6jtSr0s_tZqhheg9RAHA8X9]1xbet ru актуальное зеркало рабочее на сегодня сейчас stavka2021 ru мобильная версия скачать на андроид 1хбет букмекерская контора russian ru 2022[/url]
[url=https://vk.com/1x_zerkalo_rabotayuschee]Зеркало 1xbet работающее на сегодня прямо сейчас[/url]
https://vk.com/zerkalo_registraciya_1x
Reginaemeni
Здрасте согласен и даже более Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=oIjz8qW51f0 https://www.youtube.com/watch?v=xi1hwIN1bUM https://www.youtube.com/watch?v=Pq5Lv3F7Ifc https://www.youtube.com/watch?v=WFmOB-PLpAY https://www.youtube.com/watch?v=awnPK73IUEQ https://www.youtube.com/watch?v=Yua4RELtFCQ
[url=https://www.youtube.com/watch?v=awnPK73IUEQ]Как скачать 1хбет зеркало на айфон - 1xbet на андроид рабочее на сегодня 2022[/url] [url=https://www.youtube.com/watch?v=cnFb5JF9I74]1xbet зеркало рабочее на сегодня прямо сейчас мобильная версия скачать айфон в онлайне 1хбет[/url [url=https://www.youtube.com/watch?v=Yo9WounjURA]Где взять 1xbet промокод при регистрации где смотреть если уже зарегистрирован 1хбет за регистрацию[/url] [url=https://www.youtube.com/watch?v=2VibXiXfzCo]Регистрация 1XBET промокод при регистрации 1хбет на бесплатную ставку 2022[/url] [url=https://www.youtube.com/watch?v=a4K_s3jd5sA]Как скачать 1хбет зеркало на айфон бесплатно 1xbet на андроид на русском зеркало #Shorts 2022[/url] [url=https://www.youtube.com/watch?v=HLrq-tjYTx8]Промокод 1XBET при регистрации на деньги 1хбет регистрация Промокод бесплатно Казахстан как получить[/url]
https://www.youtube.com/playlist?list=PLn302bf7FvWWvWLP1ieXUAcV3kaq10tQa https://www.youtube.com/playlist?list=PLmWIHsWIXHM92nM-MmO0YWUAKuxf_mdYB https://www.youtube.com/playlist?list=PLRtDAAJCNRQHSto06mBFDvBPyckWmSRoK https://www.youtube.com/playlist?list=PLSbnX-iWTiNGWcsLvWk1ytt12JA_UHoHR https://www.youtube.com/playlist?list=PLVXbMH_gmerWir4S2a4Ix4iVejt9sRZL_ https://www.youtube.com/playlist?list=PLC0Ox2GUooS81oCYy3UjU7VGM0fwtzdI- https://www.youtube.com/playlist?list=PLqar3JZGyXqtNTypu4M0KLht_H_Chogmi https://www.youtube.com/playlist?list=PLNTGp_dwgBIZF0nd6bUfBIWzC8K37Nvgf https://www.youtube.com/playlist?list=PL9P3M_2TfQen3XpxOEqUHy3cKS9GpX-HW
[url=https://www.youtube.com/playlist?list=PLn302bf7FvWWvWLP1ieXUAcV3kaq10tQa]Рабочее зеркало 1xbet мобильная версия на сегодня прямо сейчас войти в личный кабинет приложение скачать на май 2022 1хбет[/url] [url=https://www.youtube.com/playlist?list=PLmWIHsWIXHM92nM-MmO0YWUAKuxf_mdYB]Скачать 1xbet на андроид последняя мобильная версия сайта актуальное зеркало рабочее на айфон 7 ios бесплатно букмекерская контора 1хбет май 2022[/url] [url=https://www.youtube.com/playlist?list=PLRtDAAJCNRQHSto06mBFDvBPyckWmSRoK]1xbet зеркало рабочее на сегодня вход сайт 1хбет казино по смс срочно скачать через вк 2022 [/url] [url=https://www.youtube.com/playlist?list=PLSbnX-iWTiNGWcsLvWk1ytt12JA_UHoHR]1xBET бонус Промокод 1хбет 2022 Как использовать Промокод 1x Bet на бесплатную ставку 6500 Как активировать и куда вводить Лучший метод 2022[/url] [url=https://www.youtube.com/playlist?list=PLVXbMH_gmerWir4S2a4Ix4iVejt9sRZL_]1xbet рабочее зеркало на сегодня прямо сейчас 1хбет 2022[/url] [url=https://www.youtube.com/playlist?list=PLERltIqyCrX1bHEZM7SUABCc2o5Rijis0]Бесплатный промокод 1xbet Бесплатную ставку на 1хбет Бонус за регистрацию Промокод 1xbet на 6500 рублей все инструкции на сегодня Бесплатный фрибет[/url]
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url]
https://vk.com/dostup_k_saitu
KevinMix
Hello! [url=http://uffcialis.online/]order cialis pills[/url] beneficial internet site http://uffcialis.online
Kelle
I am really delighted to read this website posts which contains tons of valuable data, thanks for providing these statistics.
Reginaemeni
Здравствуйте Приятно найти единомыщленников Так вот
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=6_Tf-kz_Ljs https://www.youtube.com/watch?v=6LYHvzd3xwM https://www.youtube.com/watch?v=vH7GkC1L3po https://www.youtube.com/watch?v=Rr47JoUXnQM https://www.youtube.com/watch?v=LnMDBErfRnA https://www.youtube.com/watch?v=cgZRqrf84KM
[url=https://www.youtube.com/watch?v=nAUbLZOMfU8]1XBET регистрация по телефону и вход в личный кабинет как зарегистрироваться по номеру телефона[/url] [url=https://www.youtube.com/watch?v=MJhG3LuCWTw]1xbet регистрация Казахстан вход в личный кабинет Казакша вход в личный кабинет в 1хбет - скачать[/url] [url=https://www.youtube.com/watch?v=ZGiZvXquPcQ]1xbet регистрация килиш по номеру телефона для компьютера личный кабинет[/url] [url=https://www.youtube.com/watch?v=g_d0Zyl2VqE]1XBET зеркало регистрация в 1 клик по номеру телефона в один клик вход 1ХБЕТ[/url] [url=https://www.youtube.com/watch?v=vIZD9oOl0tg]1XBET зеркало регистрация в 1 клик по номеру телефона в один клик вход 1ХБЕТ[/url] [url=https://www.youtube.com/watch?v=TMBWt3Xf5xw]1xbet регистрация промокод при регистрации 1хбет 2022 [/url] [url=https://www.youtube.com/watch?v=fieWXlMi01o]1xbet ?алай регистрация жасайды іспит иностранных граждан в России[/url] [url=https://www.youtube.com/watch?v=GhBNvgzg5oc]Как узнать свой промокод 1xbet как добавить промо код в 1хбет на 2022 бк[/url]
https://www.youtube.com/playlist?list=PLlczAilWsh0Nq49NxuCqOqIVEKEo3XGmn https://www.youtube.com/playlist?list=PLUtueTjtNu2wPwG53EEZKh_q7p22xKN6P https://www.youtube.com/playlist?list=PLKksAEBInoMBAXrqDemApw5GdRrgqL-DA https://www.youtube.com/playlist?list=PL4CjRn6RdKsBvVbUctLCwBJ7lkOzEt7k- https://www.youtube.com/playlist?list=PLvZ0_MYm3Pnbc7XGOTnT-dCH_PHH1LUAx https://www.youtube.com/playlist?list=PLO5fmvFT3pPrCnoKra8wQj4qXe7gVJtgU
[url=https://www.youtube.com/playlist?list=PLM96uXdkDUMX4E09kVqfx3gK5eiLUIEE]3 бесплатных колеса в 1Xbet бесплатная ставка в 1хбет промокод на 32500 тысячи рублей Квесты дня КАК ПОЛУЧИТЬ БЕСПЛАТНЫЕ ДЕНЬГИ НА 1XBET в 2022г[/url] [url=https://www.youtube.com/playlist?list=PLc-90dyP3JwjLzvdTyL2ifUoxqPZZP1ab]What is lamba 1xbet promo code 2022 bangladesh mc lively 1x bet promocode kazakhstan[/url] [url=https://www.youtube.com/playlist?list=PLOpri_vcHMqVEQ9V0Iw7lBJG27z3MjFgy]1xBET бонус Промокод 1хбет 2022 Как использовать Промокод 1x Bet на бесплатную ставку 6500 Как активировать и куда вводить Лучший метод 2022[/url] [url=https://www.youtube.com/playlist?list=PLF3Sux-R2AuPB641dStr6hmkbKn0hiQMd]Смотреть полный обзор конторы 1XBET Узбекистан 2022 Использовать промокод 1хбет на бонус при регистрации 1x Bet Куда и как вводить промокод 1х Бет[/url] [url=https://www.youtube.com/playlist?list=PLq4BI-2vvhBJyfTLSbLEWqZRFOLE95BTp]What is the meaning of promo code in 1xbet malaysia davido promocode for 1x bet maroc[/url] [url=https://www.youtube.com/playlist?list=PLq4BI-2vvhBIt-hdW9B38vkjjlCUw3OaZ]Exemple de code promo 1xbet pour parier gratuitement free spins no deposit pakistan gift[/url] [url=https://www.youtube.com/playlist?list=PLq4BI-2vvhBItz099YXZM6zadBm_2H332]Sydney promo code for free bet in 1xbet how do i get my 1x bet promocode iran gift Casino[/url] [url=https://www.youtube.com/playlist?list=PLq4BI-2vvhBLd49W0EdauNt2qreeVKpVu]What is the promo code for 1xbet in zambia how to get promocode in 1 xbet lord zeus[/url] [url=https://www.youtube.com/playlist?list=PLq4BI-2vvhBJs9ahaD-PCUn5-GAkFsENn]Code promo pour parier gratuitement sur 1xbet a quoi sert le code promo 1x bet qatar 2022[/url] [url=https://www.youtube.com/playlist?list=PLq4BI-2vvhBJPbLGJE3v-YOxtje8e5MT]Comment creer mon propre code promo 1xbet quel est le code promo sur 1x bet qatar telegram[/url] [url=https://www.youtube.com/playlist?list=PLq4BI-2vvhBISpMDCDBsRv-Tq2ZmOJNme]Comment parier avec un code promo 1xbet c’est quoi le code promo 1xbet quran philippines[/url]
[url=https://vk.com/zerkalo_registraciya_1x]ЗЕРКАЛО 1xbet регистрация на сайте в один клик[/url]
https://vk.com/1x_zerkalo_rabotayuschee
Reginaemeni
Здравствуйте хорошая мысль но Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=ZGiZvXquPcQ https://www.youtube.com/watch?v=g_d0Zyl2VqE https://www.youtube.com/watch?v=vIZD9oOl0tg https://www.youtube.com/watch?v=HLrq-tjYTx8 https://www.youtube.com/watch?v=TMBWt3Xf5xw https://www.youtube.com/watch?v=fieWXlMi01o https://www.youtube.com/watch?v=GhBNvgzg5oc
[url=https://www.youtube.com/watch?v=tqrBVSmvglM]Как создать свой промокод 1xbet как сделать промо код в 1хбет[/url] [url=https://www.youtube.com/watch?v=Pq5Lv3F7Ifc]1xbet обзор о конторе 1xbet промокод 2022 1хбет при регистрации на деньги что дает 2022[/url] [url=https://www.youtube.com/watch?v=wxyLpD-Qfu8]Как получить промокод на бесплатную ставку 1xbet промокоды на бесплатные вращения 1хбет 2022[/url] [url=https://www.youtube.com/watch?v=oIjz8qW51f0]Как использоватьпромокод на бесплатную ставку 1xbet бездепозитный бонус бесплатно 1хбет 2022[/url] [url=https://www.youtube.com/watch?v=xi1hwIN1bUM]1xбет промокод при регистрации казахстан инструкция казакша 1хбет калай жазады 2022[/url] [url=https://www.youtube.com/watch?v=WFmOB-PLpAY]1xbet ЗЕРКАЛО рабочее на сегодня прямо сейчас скачать актуальное на айфон май 2022 1хбет[/url] [url=https://www.youtube.com/watch?v=Yua4RELtFCQ]Как скачать 1хбет зеркало на айфон - 1xbet на андроид рабочее на сегодня 2022[/url]
https://www.youtube.com/playlist?list=PLjdCroy9jYtCzLsiYUVh5Q5kEBSkpacVR https://www.youtube.com/playlist?list=PLR1JbWGthH9a6LsQF8EWdlxOf-yjvj5Vf https://www.youtube.com/playlist?list=PLEJlwUoSx-l9N_MVOOJvGH8viprtihOIE https://www.youtube.com/playlist?list=PLXoRDvIP0JHh5xDV1UE7MsHplkEUtZYU_ https://www.youtube.com/playlist?list=PLa_Jln4KYdhFRl9nq0kOYJdBRvs15tbVk https://www.youtube.com/playlist?list=PLJpkz6b0bHKaUA_aHzcuYhMl9yI9Ahp7e https://www.youtube.com/playlist?list=PLzaZ8Pqna7gNXgq8bz92U6s0GBDhIQ8xi https://www.youtube.com/playlist?list=PLCGD4gHObORmtp6XH0qOtCq6QlmHXbRH0
[url=https://www.youtube.com/playlist?list=PLINg0JEvN_DCTARops_G9Nzhbg7osXQAW]1xbet зеркало на ios - как скачать в apple store на iphone и ipad регистрация в 1хбет на андроид ссылка мобильная версия Зеркало альтернативный вход[/url] [url=https://www.youtube.com/playlist?list=PLY0GQ_p6e4p3aqyxgpUdb8Oh5n0er4jq8]1xbet вход в личный кабинет зеркало сайта работающее Россия скачать на андроид 2022 1хбет на сегодняшний день на май месяц[/url] [url=https://www.youtube.com/playlist?list=PL5L59fToCm5WwTPunfbYOKAuButsI4IZU]Рабочее зеркало 1xbet мобильная версия на сегодня прямо сейчас войти в личный кабинет приложение скачать на май 2022 1хбет[/url] [url=https://www.youtube.com/playlist?list=PL8TUY3TWrfUgRIsczu_VYWgcbl4bA1EpX]Скачать 1XBET на АНДРОИД. Приложение 1ХБЕТ Мобильная версия ссылка зеркало скачать на андроид 2022 ссылка мобильная версия зеркало[/url] [url=https://www.youtube.com/playlist?list=PL5FufKBP9HpUrqy9Kb7w46uISoLtAgITW]Как скачать зеркало 1xbet на андроид казино зеркало мобильная версия один икс бэд май 2022[/url]
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url]
https://vk.com/zerkalo_rabochee_1x
Samara
king’s college london coursework course work hero differential equations coursework
Reginaemeni
Здравствуйте Поддерживаю Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=ZGiZvXquPcQ https://www.youtube.com/watch?v=g_d0Zyl2VqE https://www.youtube.com/watch?v=vIZD9oOl0tg https://www.youtube.com/watch?v=HLrq-tjYTx8 https://www.youtube.com/watch?v=TMBWt3Xf5xw https://www.youtube.com/watch?v=fieWXlMi01o https://www.youtube.com/watch?v=GhBNvgzg5oc
[url=https://www.youtube.com/watch?v=OGMzex2lcPk]Best promo code for 1xbet bangladesh 2022 free for registration android[/url] [url=https://www.youtube.com/watch?v=XA0GpOcbOSs]How to use accumulator promo code in 1xbet kivabe data truncated for column[/url] [url=https://www.youtube.com/watch?v=ZHhmI3sjIN4]How to use accumulator promo code in 1xbet kivabe data truncated for column[/url] [url=https://www.youtube.com/watch?v=PrCOCN3b9IA]How does 1xbet promo code work india registration bonus casino 2022 [/url] [url=https://www.youtube.com/watch?v=5eH41BAc9LY]How does 1xbet promo code work india registration bonus casino 2022 [/url] [url=https://www.youtube.com/watch?v=Hqk5sQSyFnE]1xbet promo code 2022 Somali comment parier avec un code promo sur 1xbet[/url]
https://www.youtube.com/playlist?list=PLPPJ1hx14qAhlmppprSGnjJhe3krNF9SK https://www.youtube.com/playlist?list=PLQI1Vc8W_P6jtSr0s_tZqhheg9RAHA8X9 https://www.youtube.com/playlist?list=PL_Fg7H0B2VyQ3ENndJNKz-SoYaJXi5UPO https://www.youtube.com/playlist?list=PLqX6jWtLeZXPd8q7xD3i3q2dt9t5JrQkt https://www.youtube.com/playlist?list=PL7-zQT8YROb-eD46AlzexYfi-IKLk0bkf https://www.youtube.com/playlist?list=PLiB1WY8HrjYOecPrNyYJNRoffoa63YxI8 https://www.youtube.com/playlist?list=PLAjutnTgtqg0BudmWmkM5h7h7EYHDkDe4 https://www.youtube.com/playlist?list=PL-l1a-GqnPc5FjksiKcldxZ1AZsh7O7EP
[url=https://www.youtube.com/playlist?list=PLq4BI-2vvhBJyfTLSbLEWqZRFOLE95BTp]What is the meaning of promo code in 1xbet malaysia davido promocode for 1x bet maroc[/url] [url=https://www.youtube.com/playlist?list=PLox3iLQrr_mJ_jq6Ab19Vbiqoxb2KfufP]How to check 1xbet promo code nigeria comment avoir le codepromo 1x bet generator[/url] [url=https://www.youtube.com/playlist?list=PL5kshdwDp37-EYzMGp3RODaWnKXlLQtlf]Comment avoir son code promo 1xbet how to use 1x bet free bet promocode bonus[/url] [url=https://www.youtube.com/playlist?list=PLDEo_poCDpdmH_JJgVQIKFW8yJrvKBZgc]What is davido 1xbet promo code 2022 nigeria uae how to use 1x bet gift promocode[/url] [url=https://www.youtube.com/playlist?list=PLlczAilWsh0Nq49NxuCqOqIVEKEo3XGmn]Comment parier avec un code promo 1xbet c’est quoi le code promo 1xbet quran philippines[/url] [url=https://www.youtube.com/playlist?list=PLUtueTjtNu2wPwG53EEZKh_q7p22xKN6P]Comment creer mon propre code promo 1xbet quel est le code promo sur 1x bet qatar telegram[/url] [url=https://www.youtube.com/playlist?list=PLKksAEBInoMBAXrqDemApw5GdRrgqL-DA]What is davido 1xbet promo code 2022 nigeria uae how to use 1x bet gift promocode[/url]
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url]
https://vk.com/zerkalo_rabochee_1x
Antony
Connecticut Searching for the greatest Connecticut on the internet casinos in 2022?
WideRange
The not small EV charger with built-in found in the extreme detection. We just offer EV puff types based on recommended electrical code. No more inopportune breakers tripping. Our EV charger power settings do not overwhelm the plug’s capacity.
ev chargers are here http://ev-chargers.com/kia/niro/
[url=http://ev-chargers.com/audi/audi-q5-ev-chargers/]how you can help[/url]
<a href=http://ev-chargers.com/nema-14-50/>additional hints</a>
KendraEmurf
Привет хорошая мысль но Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=ZGiZvXquPcQ https://www.youtube.com/watch?v=g_d0Zyl2VqE https://www.youtube.com/watch?v=vIZD9oOl0tg https://www.youtube.com/watch?v=HLrq-tjYTx8 https://www.youtube.com/watch?v=TMBWt3Xf5xw https://www.youtube.com/watch?v=fieWXlMi01o https://www.youtube.com/watch?v=GhBNvgzg5oc
https://www.youtube.com/playlist?list=PL2yED4GdwUDgom-z9z8TnkCh4Ks7ybBsv https://www.youtube.com/playlist?list=PL2yED4GdwUDhEaDJlEAPcA9IYMyuyh6iO https://www.youtube.com/playlist?list=PL2yED4GdwUDi7OZu0m8NSb2YwSlqc2Z3t https://www.youtube.com/playlist?list=PL2yED4GdwUDgUVnmY0wOxvXcScmgmwIJh https://www.youtube.com/playlist?list=PL2yED4GdwUDiRgXdQ7vbmjbWOTGZPmkVm https://www.youtube.com/playlist?list=PL2yED4GdwUDjcmB6aoBGBDgnQz_oqXIim https://www.youtube.com/playlist?list=PL2yED4GdwUDhqt03JAMn53M-KrNKVJyf3 https://www.youtube.com/playlist?list=PL2yED4GdwUDizKh5xAywaGDyir1xeAovf
https://www.youtube.com/playlist?list=PLq4BI-2vvhBIqLGPBPTc5ajwQLKPt3qku https://www.youtube.com/playlist?list=PLq4BI-2vvhBKPwrIEUEFeHH3KoXotByio https://www.youtube.com/playlist?list=PLq4BI-2vvhBIDVcvQLRwU3cr_gheV1qnp https://www.youtube.com/playlist?list=PLq4BI-2vvhBLoRRNWdgFMRGjxSdNaCWsC https://www.youtube.com/playlist?list=PLq4BI-2vvhBLoYurr-FLynMsXOGf7fCss https://www.youtube.com/playlist?list=PLq4BI-2vvhBKwAyyBQVZt4mRfpd2MBzF9 https://www.youtube.com/playlist?list=PLq4BI-2vvhBLrp8mENpEIsurTdBe1JxGE https://www.youtube.com/playlist?list=PLq4BI-2vvhBI3uain9fUkozCGiJUu_u3g https://www.youtube.com/playlist?list=PLq4BI-2vvhBIPuVjSapf4tMbvMls8Rpfr
Charlesjange
купить мобильный sip номер https://continent-telecom.com
KendraEmurf
Здрасте Поддерживаю Так вот
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=ZGiZvXquPcQ https://www.youtube.com/watch?v=g_d0Zyl2VqE https://www.youtube.com/watch?v=vIZD9oOl0tg https://www.youtube.com/watch?v=HLrq-tjYTx8 https://www.youtube.com/watch?v=TMBWt3Xf5xw https://www.youtube.com/watch?v=fieWXlMi01o https://www.youtube.com/watch?v=GhBNvgzg5oc
https://www.youtube.com/playlist?list=PL2yED4GdwUDgom-z9z8TnkCh4Ks7ybBsv https://www.youtube.com/playlist?list=PL2yED4GdwUDhEaDJlEAPcA9IYMyuyh6iO https://www.youtube.com/playlist?list=PL2yED4GdwUDi7OZu0m8NSb2YwSlqc2Z3t https://www.youtube.com/playlist?list=PL2yED4GdwUDgUVnmY0wOxvXcScmgmwIJh https://www.youtube.com/playlist?list=PL2yED4GdwUDiRgXdQ7vbmjbWOTGZPmkVm https://www.youtube.com/playlist?list=PL2yED4GdwUDjcmB6aoBGBDgnQz_oqXIim https://www.youtube.com/playlist?list=PL2yED4GdwUDhqt03JAMn53M-KrNKVJyf3 https://www.youtube.com/playlist?list=PL2yED4GdwUDizKh5xAywaGDyir1xeAovf
https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJiG7pECb7QDPwpDPhJlfCh https://www.youtube.com/playlist?list=PLq4BI-2vvhBJORFMgQteH3OGrZFAQRruY https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBI-Ab6axauG7f0PMciZvhxo https://www.youtube.com/playlist?list=PLq4BI-2vvhBLxpjDLTpoO1htzns24L7XQ https://www.youtube.com/playlist?list=PLq4BI-2vvhBJfSYQYyuDb9Jy7Bdz155kS https://www.youtube.com/playlist?list=PLq4BI-2vvhBIwDotoD1zozfoxAT1vioi3
Elliot
I am really inspired with your writing talents as well as with the layout in your blog. Is this a paid subject or did you customize it your self? Either way keep up the excellent high quality writing, it is uncommon to peer a great blog like this one these days..
Bridgett
Hey there! Do you use Twitter? I’d like to follow you if that would be okay. I’m undoubtedly enjoying your blog and look forward to new updates.
Wilson
I am really pleased to read this weblog posts which includes tons of useful information, thanks for providing such information.
Connie
I think this is among the most vital info for me. And i’m glad reading your article. But wanna remark on few general things, The website style is wonderful, the articles is really great : D. Good job, cheers
KendraEmurf
Салют согласен и даже более И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=cnFb5JF9I74 https://www.youtube.com/watch?v=Yo9WounjURA https://www.youtube.com/watch?v=2VibXiXfzCo https://www.youtube.com/watch?v=a4K_s3jd5sA https://www.youtube.com/watch?v=Ooe7Zhr8Ios https://www.youtube.com/watch?v=nAUbLZOMfU8 https://www.youtube.com/watch?v=MJhG3LuCWTw
https://www.youtube.com/playlist?list=PLneJ0QN_YNk6baz9zeudBhUCzYlGtPE_0 https://www.youtube.com/playlist?list=PLneJ0QN_YNk7Y8tx-VgSnl7ubzdIKCRnX https://www.youtube.com/playlist?list=PLneJ0QN_YNk5iRlC3MVh2RsAFmB88YSZE https://www.youtube.com/playlist?list=PLneJ0QN_YNk6OLDy8SaIvgWsMxEHQubOo https://www.youtube.com/playlist?list=PLneJ0QN_YNk7jFysr32Sf6Q5yu9B7eoH2 https://www.youtube.com/playlist?list=PLneJ0QN_YNk5ThXvfx3JOuKX-omxqpcHK https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBrBmHmAX_RHanGvmD464yf https://www.youtube.com/playlist?list=PLRcnUiG5ZvBAUZm9GihHfYfDK_Ftm7HoM
https://www.youtube.com/playlist?list=PLq4BI-2vvhBKpthxPvURqkFnAAusp5gxu https://www.youtube.com/playlist?list=PLq4BI-2vvhBLuFrOMIPPw8ulahnfk6X94 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJng2iNLNFPQ0l6HLq5keQD https://www.youtube.com/playlist?list=PLq4BI-2vvhBKIH5Cy18DUUAuePgHYeNXX https://www.youtube.com/playlist?list=PLq4BI-2vvhBKnv_CwIoCcDypasoI5d4Tc https://www.youtube.com/playlist?list=PLq4BI-2vvhBLMogknP6oCK5QwUwo4AfWM
KendraEmurf
Hi Приятно найти единомыщленников И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=6_Tf-kz_Ljs https://www.youtube.com/watch?v=6LYHvzd3xwM https://www.youtube.com/watch?v=vH7GkC1L3po https://www.youtube.com/watch?v=Rr47JoUXnQM https://www.youtube.com/watch?v=LnMDBErfRnA https://www.youtube.com/watch?v=cgZRqrf84KM
https://www.youtube.com/playlist?list=PLNzG2x9bf1wwlbMby1ccV9htGRMfimWQH https://www.youtube.com/playlist?list=PLNzG2x9bf1wyjCzuyPrDuDytj9WQCYorr https://www.youtube.com/playlist?list=PLneJ0QN_YNk7h8FCoR5PDaQIWe7fmrE-z https://www.youtube.com/playlist?list=PLneJ0QN_YNk7iaBy95I0GkhDaZzRjqrjc https://www.youtube.com/playlist?list=PLneJ0QN_YNk4Nlh2_RqfdJ5PTNdjQ1Go- https://www.youtube.com/playlist?list=PLneJ0QN_YNk7BkdRH73blRJAzAekfneuk https://www.youtube.com/playlist?list=PLneJ0QN_YNk4E8VmkBR9Mix92IoXC7WDK https://www.youtube.com/playlist?list=PLneJ0QN_YNk5VsHxzDYTkDhtoC0DZf2uv
https://www.youtube.com/playlist?list=PLq4BI-2vvhBKpthxPvURqkFnAAusp5gxu https://www.youtube.com/playlist?list=PLq4BI-2vvhBLuFrOMIPPw8ulahnfk6X94 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJng2iNLNFPQ0l6HLq5keQD https://www.youtube.com/playlist?list=PLq4BI-2vvhBKIH5Cy18DUUAuePgHYeNXX https://www.youtube.com/playlist?list=PLq4BI-2vvhBKnv_CwIoCcDypasoI5d4Tc https://www.youtube.com/playlist?list=PLq4BI-2vvhBLMogknP6oCK5QwUwo4AfWM
Robbin
She was inside her family’s property in Weest Valley City when she was struck.
BrendaTed
Салют согласен и даже более И вот это тоже по теме Parimatch зеркало сайта работающее всегда
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=rK5Dec32OHk https://www.youtube.com/watch?v=wNm_1hTV5Io https://www.youtube.com/watch?v=T8noWHJbRd8 https://www.youtube.com/watch?v=LVEEc8Dt0Nk https://www.youtube.com/watch?v=GWB1n6Yje24 https://www.youtube.com/watch?v=ntFrCCPkUqM https://www.youtube.com/watch?v=FfTYT7GSb_A
[url=https://www.youtube.com/watch?v=GgygicCW120]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=jI9fRdPcauM]ПРОМОКОД Париматч фриспины Parimatch на бесплатные фриспины за регистрацию Пари Матч 99 без депозита[/url] [url=https://www.youtube.com/watch?v=r_HRxxhD9oQ]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=PwH_uOJD7P8]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url]
https://www.youtube.com/playlist?list=PLnsnFM60Rk1S_Jargkz9EpKhVqjn6itat https://www.youtube.com/playlist?list=PLoV15PAdIrbtRnmM5b2UJt31V7wXVZ7G9 https://www.youtube.com/playlist?list=PLubYmKbhMVAg4jncddzEkbOMHLUf_GccA https://www.youtube.com/playlist?list=PL1R7UsD2cHeCdVrhJ6TOER4bxWQ7ApY5i https://www.youtube.com/playlist?list=PLlh1XFwgra_2pLsVUhhfouib0cwDVgb4y https://www.youtube.com/playlist?list=PLiHyRoZH_meBNtosHJRoHt6B7p1S2RiFS https://www.youtube.com/playlist?list=PLCsGp5QHVXdo58bY_AH_TZxNNqpvK0aGC https://www.youtube.com/playlist?list=PLlh1XFwgra_2pLsVUhhfouib0cwDVgb4y https://www.youtube.com/playlist?list=PLIdfnJD5haBI9MTXFuet6j3_JOY4PSWjz
[url=https://www.youtube.com/playlist?list=PLTivr89Sv4_1PUD7y4uv2Q6juTvOJutqe]Где ввести промокод Париматч Беларусь на промокоды Parimatch Украина Пари Матч КЗ 2022[/url] [url=https://www.youtube.com/playlist?list=PLV9fx-snYv-BcNVbkklhgBK39od15FTCs]Промокод на Пари Матч Казино Париматч фриспины в слоты и покер 2022 Parimatch promo code[/url] [url=https://www.youtube.com/playlist?list=PL2xeIlkrOHgCac9hUQA_LLq_vDtPMcs_c]Промокод Париматч без депозита на фрибет и фриспины казино промокоды Parimatch на бездеп[/url] [url=https://www.youtube.com/playlist?list=PLmwETSG8roNeOeyJ-2J0oyZv9AvyQCftJ]Промокод Париматч 2022 Украина Parimatch промокоды Пари Матч бонус без депозита регистрация[/url]
https://vk.com/bukmekerskaya_kontora_partch
[url=https://vk.com/bukmekerskaya_kontora_parimatch]Букмекерская контора Париматч регистрация бонус[/url]
BrendaTed
Привет Поддерживаю Так же хочу добавить Parimatch зеркало сайта работающее последнее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=t9cKRIAE3Lk https://www.youtube.com/watch?v=KfaPhR7MTrU https://www.youtube.com/watch?v=ysWQS-zwhkQ https://www.youtube.com/watch?v=fPNSj0ckYnk https://www.youtube.com/watch?v=5x8dzT0BQHA https://www.youtube.com/watch?v=tjoUGu5rGfY https://www.youtube.com/watch?v=WiPPQPfdqZ8 https://www.youtube.com/watch?v=uAgxzu33ZIk
[url=https://www.youtube.com/watch?v=k6hfH6RB9V0]Промокод на фриспины 77 Париматч с депозитом только за регистрацию 2022 Parimatch[/url] [url=https://www.youtube.com/watch?v=P4cadSLQ1rU]Париматч Украина игровые автоматы бонус за регистрацию бездепозитный на реальные деньги[/url] [url=https://www.youtube.com/watch?v=p2xEzaH0FRo]Париматч игровые автоматы минимальный депозит їх бонуси за регистрацию отзывы[/url] [url=https://www.youtube.com/watch?v=AROs8UqEujI]Париматч промокод при регистрации без депозита 2022 promo bonus Parimatch за верификацию[/url]
https://www.youtube.com/playlist?list=PLS_6GTVOYC9gH201sT1rRk8kYOTN6c_91 https://www.youtube.com/playlist?list=PLGarVr4eBmsmXt4UriVNDPOA2tQ_a2Fow https://www.youtube.com/playlist?list=PL3iExQOpO3rSoQPUXZnXmqQajfDvwyqPf https://www.youtube.com/playlist?list=PL6mBd_duFt_b3NHcudr1GV0I2xNVrc35t https://www.youtube.com/playlist?list=PLqP_K6SSpMSHwzaPnha4AhmBvQa09O50U https://www.youtube.com/playlist?list=PLW6Elf6cZK1LuxbvdHN4lgTVZruk56Iln https://www.youtube.com/playlist?list=PLW0QWsWSrepme9pUtBWZdybI_HTtVYrEJ https://www.youtube.com/playlist?list=PLqsM6sDvDQzDPoPHCFY8k0DC9QUrzvtOX https://www.youtube.com/playlist?list=PLk2Q0fXi4isJE_eAwXEdFsUMy5E-kVyDd
[url=https://www.youtube.com/playlist?list=PL1D6Led19hjDG61mPEC8Wii-AKMHOCtfO]Промокод БОНУС Париматч Слоты Которые Дают 2022 самые выигрышные дающие с бонусами популярные слот в Parimatch[/url] [url=https://www.youtube.com/playlist?list=PLBLO37Hy6RNAUMgYwQErjaDuI-EAlDfdE]Промокод БОНУС Париматч Слоты Которые Дают 2022 самые выигрышные дающие с бонусами популярные слот в Parimatc Украина детальный обзор отзывы[/url] [url=https://www.youtube.com/playlist?list=PLN5HOHzLUbahEWq8YAM8WhD-4Y58ANPp-]Париматч бонус за верификацию - Пари Матч бонусы при регистрации - как забрать бонус в Parimatch[/url] [url=https://www.youtube.com/playlist?list=PL90A0HJl0Q_VSkHt71-0_JYKPU4CDYCqL]Обзор онлайн Париматч контора Parimatch бонус - Промокод Париматч Как получить фриспины и бонусы в Пари Матч без депозита[/url] [url=https://www.youtube.com/playlist?list=PLtuKjXLNTK35Hk1mJ0DnsBS9W2wkm5TXe]Промокод Париматч 2022 Украина - Как зарегистрироваться на Пари-Матч и получить бонус[/url]
https://vk.com/parimatch_vhod_registraciya
[url=https://vk.com/parimatch_akciya_bonus_promokod]Париматч АКЦИИ БОНУС при регистрации промокод[/url]
Paulette
Howdy just wanted to give you a quick heads up. The words in your post seem to be running off the screen in Chrome. I’m not sure if this is a format issue or something to do with web browser compatibility but I figured I’d post to let you know. The style and design look great though!
Hope you get the issue fixed soon. Thanks
BrendaTed
Салют хорошая мысль но Так же хочу добавить Parimatch зеркало сайта работающее последнее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=t9cKRIAE3Lk https://www.youtube.com/watch?v=KfaPhR7MTrU https://www.youtube.com/watch?v=ysWQS-zwhkQ https://www.youtube.com/watch?v=fPNSj0ckYnk https://www.youtube.com/watch?v=5x8dzT0BQHA https://www.youtube.com/watch?v=tjoUGu5rGfY https://www.youtube.com/watch?v=WiPPQPfdqZ8 https://www.youtube.com/watch?v=uAgxzu33ZIk
[url=https://www.youtube.com/watch?v=eTqXSLboJ84]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=p9hG7kHsZMY]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=RQ5XYAfSshk]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=x3bRRbcN6Bo]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url]
https://www.youtube.com/playlist?list=PLlOP0S2i0zJRMv9A_8exWBE26U0pOetyM https://www.youtube.com/playlist?list=PL-oOWrKcXD00DmJbVd5SbliXo3sJR0mPn https://www.youtube.com/playlist?list=PLlprnaVTmuxLRGNtRWW8UInuNku6eMT6j https://www.youtube.com/playlist?list=PLD8Bx5tfu2pB8o1G2GsdrgTsq8OGxtAEd https://www.youtube.com/playlist?list=PLx1-Xp1bcravZrD9iaeb5St48x5Bv5uhe https://www.youtube.com/playlist?list=PL8XebQPlV77AXeikiiql_fIyBE234KH-B https://www.youtube.com/playlist?list=PLCOaM6CLKyZ861-RCfEX5MTkE4u4R0Wnc https://www.youtube.com/playlist?list=PLP14Wt7nBIGNY-5hikQzMdhg4vBi5Pr8- https://www.youtube.com/playlist?list=PLw09JdsboSXeVRhQhElN9Rs19lrlwpEIr
[url=https://www.youtube.com/playlist?list=PLlOP0S2i0zJRMv9A_8exWBE26U0pOetyM]Париматч промокод при регистрации без депозита 2022[/url] [url=https://www.youtube.com/playlist?list=PL-oOWrKcXD00DmJbVd5SbliXo3sJR0mPn]ПРОМОКОД ПАРИМАТЧ 2022 украина бездепозитный бонус[/url] [url=https://www.youtube.com/playlist?list=PLlprnaVTmuxLRGNtRWW8UInuNku6eMT6j]Промокод Париматч без депозита на фрибет и фриспины казино промокоды Parimatch на бездеп[/url]
https://vk.com/bk_pari_match_zerkalo_vhod
[url=https://vk.com/bukmekerskaya_kontora_parimatch]Париматч зеркало бк все о букмекерских конторах[/url]
Esther
Товары для животных
Nannie
My brother suggested I might like this website. He was entirely right. This post truly made my day. You can not imagine simply how much time I had spent for this information! Thanks!
Catharine
Aw, this was an incredibly good post. Taking the time and actual effort to make a good article… but what can I say… I put things off a lot and don’t seem to get anything done.
Jimmy
I seriously love your blog.. Very nice colors & theme. Did you create this amazing site yourself?
Please reply back as I’m trying to create my own personal blog and would like to learn where you got this from or exactly what the theme is named. Cheers!
Ruth
Hi there, You’ve done a great job. I will definitely digg it and personally recommend to my friends. I am confident they’ll be benefited from this website.
Alphonse
Great web site you have here.. It’s hard to find quality writing like yours nowadays. I truly appreciate individuals like you! Take care!!
Merlin
This is really interesting, You are a very skilled blogger.
I have joined your rss feed and look forward to seeking more of your fantastic post. Also, I have shared your website in my social networks!
Therese
We are a group of volunteers and starting a new scheme in our community.
Your site offered us with valuable info to work on. You’ve done an impressive job and our entire community will be thankful to you.
Rachele
I used to be able to find good advice from your articles.
AnthonyDrype
cost of stromectol medication buy stromectol online https://prednisonepills.site/# buy prednisone without prescription paypal
BrendaTed
Hi Поддерживаю Так же хочу добавить Parimatch зеркало сайта работающее последнее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=GgygicCW120 https://www.youtube.com/watch?v=jI9fRdPcauM https://www.youtube.com/watch?v=r_HRxxhD9oQ https://www.youtube.com/watch?v=PwH_uOJD7P8 https://www.youtube.com/watch?v=CU6eili0m0g https://www.youtube.com/watch?v=oxbcvcGd91w https://www.youtube.com/watch?v=yDfhfQGaKHQ https://www.youtube.com/watch?v=je1OM3vzLPI https://www.youtube.com/watch?v=ZtLOh64g6Zo
[url=https://www.youtube.com/watch?v=KfaPhR7MTrU]Промокод Париматч 2022 Украина за регистрацию Parimatch промокоды Пари Матч бонус без депозита[/url] [url=https://www.youtube.com/watch?v=t9cKRIAE3Lk]Промокод Париматч 2022 Украина за регистрацию Parimatch промокоды Пари Матч бонус без депозита[/url] [url=https://www.youtube.com/watch?v=ysWQS-zwhkQ]Промокод Париматч 2022 Украина за регистрацию Parimatch промокоды Пари Матч бонус без депозита[/url] [url=https://www.youtube.com/watch?v=fPNSj0ckYnk]Промокод Париматч 2022 Украина за регистрацию Parimatch промокоды Пари Матч бонус без депозита[/url]
https://www.youtube.com/playlist?list=PLlOP0S2i0zJRMv9A_8exWBE26U0pOetyM https://www.youtube.com/playlist?list=PL-oOWrKcXD00DmJbVd5SbliXo3sJR0mPn https://www.youtube.com/playlist?list=PLlprnaVTmuxLRGNtRWW8UInuNku6eMT6j https://www.youtube.com/playlist?list=PLD8Bx5tfu2pB8o1G2GsdrgTsq8OGxtAEd https://www.youtube.com/playlist?list=PLx1-Xp1bcravZrD9iaeb5St48x5Bv5uhe https://www.youtube.com/playlist?list=PL8XebQPlV77AXeikiiql_fIyBE234KH-B https://www.youtube.com/playlist?list=PLCOaM6CLKyZ861-RCfEX5MTkE4u4R0Wnc https://www.youtube.com/playlist?list=PLP14Wt7nBIGNY-5hikQzMdhg4vBi5Pr8- https://www.youtube.com/playlist?list=PLw09JdsboSXeVRhQhElN9Rs19lrlwpEIr
[url=https://www.youtube.com/playlist?list=PLlh1XFwgra_2pLsVUhhfouib0cwDVgb4y]Париматч как отыграть бонус - как получить бонус за регистрацию Пари Матч - где взять промокод на Parimatch[/url] [url=https://www.youtube.com/playlist?list=PLiHyRoZH_meBNtosHJRoHt6B7p1S2RiFS]Промокод Париматч без депозита - Казино Париматч регистрация, вывод денег, отзывы, бонусы, слоты, официальный сайт[/url] [url=https://www.youtube.com/playlist?list=PLCsGp5QHVXdo58bY_AH_TZxNNqpvK0aGC]Parimatch бонусы за верификацию - Бесплатные спины Пари Матч Париматч промокод Как получить бонус в париматч?[/url] [url=https://www.youtube.com/playlist?list=PLlh1XFwgra_2pLsVUhhfouib0cwDVgb4y]Париматч как отыграть бонус - как получить бонус за регистрацию Пари Матч - где взять промокод на Parimatch[/url] [url=https://www.youtube.com/playlist?list=PLIdfnJD5haBI9MTXFuet6j3_JOY4PSWjz]Parimatch промокод - Париматч Регистрация 2022 с бонусом 2500 Как получить бонус Пари Матч при Регистрации в 2022[/url]
https://vk.com/parimatch_zerkalo_rabohcee_2021
[url=https://vk.com/bk_pari_match_zerkalo_vhod]БК Пари Матч зеркало входа на сайт работающее 2022[/url]
BrendaTed
Hi со свем согласен Вот что я хочу добавить Parimatch зеркало последнее работающее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=GgygicCW120 https://www.youtube.com/watch?v=jI9fRdPcauM https://www.youtube.com/watch?v=r_HRxxhD9oQ https://www.youtube.com/watch?v=PwH_uOJD7P8 https://www.youtube.com/watch?v=CU6eili0m0g https://www.youtube.com/watch?v=oxbcvcGd91w https://www.youtube.com/watch?v=yDfhfQGaKHQ https://www.youtube.com/watch?v=je1OM3vzLPI https://www.youtube.com/watch?v=ZtLOh64g6Zo
[url=https://www.youtube.com/watch?v=GgygicCW120]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=jI9fRdPcauM]ПРОМОКОД Париматч фриспины Parimatch на бесплатные фриспины за регистрацию Пари Матч 99 без депозита[/url] [url=https://www.youtube.com/watch?v=r_HRxxhD9oQ]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url] [url=https://www.youtube.com/watch?v=PwH_uOJD7P8]Париматч промокод при регистрации бонус без депозита 2022 promo bonus Parimatch за верификацию[/url]
https://www.youtube.com/playlist?list=PL1D6Led19hjDG61mPEC8Wii-AKMHOCtfO https://www.youtube.com/playlist?list=PLBLO37Hy6RNAUMgYwQErjaDuI-EAlDfdE https://www.youtube.com/playlist?list=PLN5HOHzLUbahEWq8YAM8WhD-4Y58ANPp- https://www.youtube.com/playlist?list=PL90A0HJl0Q_VSkHt71-0_JYKPU4CDYCqL https://www.youtube.com/playlist?list=PLtuKjXLNTK35Hk1mJ0DnsBS9W2wkm5TXe https://www.youtube.com/playlist?list=PLYU7T1QT5d_P04Rc4csQNgOq4XZn2J3UQ https://www.youtube.com/playlist?list=PLZNC0fOGfVEi2ncf0aX9f-DbSDIkF-lYi https://www.youtube.com/playlist?list=PLvwjeYnP3Hn5hz9bbijf_DeHqWFJiZ-2K https://www.youtube.com/playlist?list=PLgmA4Qx-NO-R4lIJU7Wp5x_RpLAfW7m9M
[url=https://www.youtube.com/playlist?list=PLYU7T1QT5d_P04Rc4csQNgOq4XZn2J3UQ]Где ввести промокод Париматч Беларусь на промокоды Parimatch Украина Пари Матч КЗ 2022[/url] [url=https://www.youtube.com/playlist?list=PLZNC0fOGfVEi2ncf0aX9f-DbSDIkF-lYi]Пари Матч Промокод при регистрации - Обзор казино Париматч Слоты, Бонусы, Регистрация, Вход в Parimatch Casino[/url] [url=https://www.youtube.com/playlist?list=PLvwjeYnP3Hn5hz9bbijf_DeHqWFJiZ-2K]Пари Матч промокод фриспины Обзор Онлайн Казино Париматч Parimatch Букмекерская Контора Париматч Бонус[/url] [url=https://www.youtube.com/playlist?list=PLgmA4Qx-NO-R4lIJU7Wp5x_RpLAfW7m9M]Промокод Париматч 2022 Украина Parimatch промокоды Пари Матч бонус без депозита регистрация[/url]
https://vk.com/pari_match_stavki_na_spokt
[url=https://vk.com/bukmekerskaya_kontora_parimatch]Париматч зеркало бк все о букмекерских конторах[/url]
Monroe
It is the best time to make some plans for the future and it’s time to be happy. I have learn this post and if I may I desire to suggest you some interesting issues or tips. Maybe you can write subsequent articles referring to this article. I desire to read even more things approximately it!
Larhonda
A Los Angeles citizenship lawyer can help stroll you through the alternative ways citizenship can be acquired.
BrendaTed
Здрасте со свем согласен и даже более Вот что я хочу добавить Parimatch ком зеркало всегда работает
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=t9cKRIAE3Lk https://www.youtube.com/watch?v=KfaPhR7MTrU https://www.youtube.com/watch?v=ysWQS-zwhkQ https://www.youtube.com/watch?v=fPNSj0ckYnk https://www.youtube.com/watch?v=5x8dzT0BQHA https://www.youtube.com/watch?v=tjoUGu5rGfY https://www.youtube.com/watch?v=WiPPQPfdqZ8 https://www.youtube.com/watch?v=uAgxzu33ZIk
[url=https://www.youtube.com/watch?v=FWZqhvCT8Vk]Промокод на Пари Матч Казино Париматч фриспины в слоты и покер 2022 Parimatch promo code[/url] [url=https://www.youtube.com/watch?v=qg5QzwGZYdY]Промокод на Пари Матч Казино Париматч фриспины в слоты и покер 2022 Parimatch promo code[/url] [url=https://www.youtube.com/watch?v=Fka1Vy61sgg]Промокод на Пари Матч Казино Париматч фриспины в слоты и покер 2022 Parimatch promo code[/url] [url=https://www.youtube.com/watch?v=q8FBllok5Zg]Промокод на Пари Матч Казино Париматч фриспины в слоты и покер 2022 Parimatch promo code[/url] [url=https://www.youtube.com/watch?v=SDAYlhQL1aQ]Промокод Париматч без депозита на фрибет как получить фриспины казино промокоды Parimatch на бездеп[/url]
https://www.youtube.com/playlist?list=PLnsnFM60Rk1S_Jargkz9EpKhVqjn6itat https://www.youtube.com/playlist?list=PLoV15PAdIrbtRnmM5b2UJt31V7wXVZ7G9 https://www.youtube.com/playlist?list=PLubYmKbhMVAg4jncddzEkbOMHLUf_GccA https://www.youtube.com/playlist?list=PL1R7UsD2cHeCdVrhJ6TOER4bxWQ7ApY5i https://www.youtube.com/playlist?list=PLlh1XFwgra_2pLsVUhhfouib0cwDVgb4y https://www.youtube.com/playlist?list=PLiHyRoZH_meBNtosHJRoHt6B7p1S2RiFS https://www.youtube.com/playlist?list=PLCsGp5QHVXdo58bY_AH_TZxNNqpvK0aGC https://www.youtube.com/playlist?list=PLlh1XFwgra_2pLsVUhhfouib0cwDVgb4y https://www.youtube.com/playlist?list=PLIdfnJD5haBI9MTXFuet6j3_JOY4PSWjz
[url=https://www.youtube.com/playlist?list=PLlOP0S2i0zJRMv9A_8exWBE26U0pOetyM]Париматч промокод при регистрации без депозита 2022[/url] [url=https://www.youtube.com/playlist?list=PL-oOWrKcXD00DmJbVd5SbliXo3sJR0mPn]ПРОМОКОД ПАРИМАТЧ 2022 украина бездепозитный бонус[/url] [url=https://www.youtube.com/playlist?list=PLlprnaVTmuxLRGNtRWW8UInuNku6eMT6j]Промокод Париматч без депозита на фрибет и фриспины казино промокоды Parimatch на бездеп[/url]
https://vk.com/parimatch_poker_otzyvy_igrokov
[url=https://vk.com/bukmekerskaya_kontora_parimatch]Париматч зеркало бк все о букмекерских конторах[/url]
BrendaTed
Салют согласен со всем Так же хочу добавить Parimatch зеркало сайта работающее последнее
зеркало Parimatch - http://bit.ly/2vYi9s4
https://www.youtube.com/watch?v=eTqXSLboJ84 https://www.youtube.com/watch?v=p9hG7kHsZMY https://www.youtube.com/watch?v=RQ5XYAfSshk https://www.youtube.com/watch?v=x3bRRbcN6Bo https://www.youtube.com/watch?v=8sRWlR0ONWc https://www.youtube.com/watch?v=-JZ8I0ez0Ys https://www.youtube.com/watch?v=fSPjPMfoAoU https://www.youtube.com/watch?v=f1UnqBZXqUg
[url=https://www.youtube.com/watch?v=8sRWlR0ONWc]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=-JZ8I0ez0Ys]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=fSPjPMfoAoU]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url] [url=https://www.youtube.com/watch?v=f1UnqBZXqUg]ПРОМОКОД Parimatch на бесплатные фриспины Париматч фриспин за регистрацию Пари Матч[/url]
https://www.youtube.com/playlist?list=PLlOP0S2i0zJRMv9A_8exWBE26U0pOetyM https://www.youtube.com/playlist?list=PL-oOWrKcXD00DmJbVd5SbliXo3sJR0mPn https://www.youtube.com/playlist?list=PLlprnaVTmuxLRGNtRWW8UInuNku6eMT6j https://www.youtube.com/playlist?list=PLD8Bx5tfu2pB8o1G2GsdrgTsq8OGxtAEd https://www.youtube.com/playlist?list=PLx1-Xp1bcravZrD9iaeb5St48x5Bv5uhe https://www.youtube.com/playlist?list=PL8XebQPlV77AXeikiiql_fIyBE234KH-B https://www.youtube.com/playlist?list=PLCOaM6CLKyZ861-RCfEX5MTkE4u4R0Wnc https://www.youtube.com/playlist?list=PLP14Wt7nBIGNY-5hikQzMdhg4vBi5Pr8- https://www.youtube.com/playlist?list=PLw09JdsboSXeVRhQhElN9Rs19lrlwpEIr
[url=https://www.youtube.com/playlist?list=PLnsnFM60Rk1S_Jargkz9EpKhVqjn6itat]Промокод на Пари Матч Казино Париматч фриспины в слоты и покер 2022 Parimatch promo code[/url] [url=https://www.youtube.com/playlist?list=PLoV15PAdIrbtRnmM5b2UJt31V7wXVZ7G9]Где ввести промокод Париматч Беларусь на промокоды Parimatch Украина Пари Матч КЗ 2022[/url] [url=https://www.youtube.com/playlist?list=PLubYmKbhMVAg4jncddzEkbOMHLUf_GccA]Промокод Париматч 2022 Украина Parimatch промокоды Пари Матч бонус без депозита регистрация[/url] [url=https://www.youtube.com/playlist?list=PL1R7UsD2cHeCdVrhJ6TOER4bxWQ7ApY5i]ПРОМОКОД ПАРИМАТЧ казино 2022 - Промо код Parimatch Как получить фриспины и бонусы в Пари Матч без депозита[/url]
https://vk.com/bukmekerskaya_kontora_partch
[url=https://vk.com/bukmekerskaya_kontora_parimatch]Париматч зеркало бк все о букмекерских конторах[/url]
DorisBot
Салют согласен со всем Так вот
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=03zm0jxV1cg https://www.youtube.com/watch?v=zz5KMXa9wRc https://www.youtube.com/watch?v=gPRwGnEDsRY https://www.youtube.com/watch?v=yGH844DRB4E https://www.youtube.com/watch?v=e8JMjCBBCxA https://www.youtube.com/watch?v=Kd-9SAaRDTs https://www.youtube.com/watch?v=i6BW2pQ_vOE https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8
[url=https://www.youtube.com/watch?v=d2Lo0oQdU4s]How to see our promo code in 1xbet - how to use another promo code in 1xbet[/url] [url=https://www.youtube.com/watch?v=uQrqGrcOA68]How to get promo code in 1xbet telugu - how to generate 1xbet promo code[/url] [url=https://www.youtube.com/watch?v=Cy1SDz7HHq4]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url] [url=https://www.youtube.com/watch?v=gzYfo2U8Bko]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url] [url=https://www.youtube.com/watch?v=z68_ZsspLSU]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url]
[url=https://www.youtube.com/playlist?list=PLdX_HbqGQFWSshTXjoVBUUD91sKgkAsip]Как установить 1xbet зеркало скачать на айфон 5s бесплатно рабочее на сегодня 1хбет 2022[/url] [url=https://www.youtube.com/playlist?list=PLiu2X3QBr87zhKzyQwmB_Cpe9HyP1TjYY]1xbet актуальное зеркало на сегодняшний день сегодня прямо сейчас рабочее регистрация зеркало 1хбет 2022[/url] [url=https://www.youtube.com/playlist?list=PLqqnMYzfeDg4QD6HmHQ5UgYEUan4anEM8]1xbet зеркало сайта вход в личный кабинет рабочее на сегодня прямо сейчас 1хбет зеркало 2022[/url]
https://www.youtube.com/playlist?list=PLdX_HbqGQFWSshTXjoVBUUD91sKgkAsip https://www.youtube.com/playlist?list=PLiu2X3QBr87zhKzyQwmB_Cpe9HyP1TjYY https://www.youtube.com/playlist?list=PLqqnMYzfeDg4QD6HmHQ5UgYEUan4anEM8 https://www.youtube.com/playlist?list=PLjAsegiMOATkuJkwXY7tJGAznx3cNuOf4 https://www.youtube.com/playlist?list=PLvibuTWUKNStxbPddi1QGFkQd80pRQoud https://www.youtube.com/playlist?list=PLEYHnJIeTThw4XoVG2i6bY72O-tJh2lGI https://www.youtube.com/playlist?list=PLaHYt0vf74pUKG_BBQr4usrKkMcL2Lva0 https://www.youtube.com/playlist?list=PLFGhmqZCCaybBqMlHJD09-wPZ4Akv5CZ-
[url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня[/url] [url=https://vk.com/bet_bonus_perviy_depozit]1хбет бонус на первый депозит 100% 1xbet условия[/url] [url=https://vk.com/zerkalo__vhod_alternativniy_sait]1xbet зеркало рабочее вход альтернативный сайт[/url] https://vk.com/dostup_k_saitu https://vk.com/bet_bonus_perviy_depozit
Jamesmib
https://zithromaxpills.store/# zithromax z-pak price without insurance
DorisBot
Hi со свем согласен и даже более И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=OGMzex2lcPk https://www.youtube.com/watch?v=XA0GpOcbOSs https://www.youtube.com/watch?v=ZHhmI3sjIN4 https://www.youtube.com/watch?v=PrCOCN3b9IA https://www.youtube.com/watch?v=5eH41BAc9LY https://www.youtube.com/watch?v=Hqk5sQSyFnE https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8
[url=https://www.youtube.com/watch?v=7sTlCqagP90]How to use free bet promo code in 1xbet accumulator - 1xbet no risk bet bangla[/url] [url=https://www.youtube.com/watch?v=8c7sSUXYe_M]How to use free bet promo code in 1xbet accumulator - 1xbet no risk bet bangla[/url] [url=https://www.youtube.com/watch?v=RXnggiHXYO4]1XBET free bet promo code use - no risk bet 1XBET hindi 2022 today[/url] [url=https://www.youtube.com/watch?v=s1dwJtOs9Ek]1xbet promo codes in Pakistan 2022 Hindi Nepal new code promo 1xbet 2022 India[/url]
[url=https://www.youtube.com/playlist?list=PLjAsegiMOATkuJkwXY7tJGAznx3cNuOf4]1XBET зеркало рабочий домен на сегодня сейчас 1ХБЕТ работающий адрес сайта скачать ios зеркало 2022[/url] [url=https://www.youtube.com/playlist?list=PLvibuTWUKNStxbPddi1QGFkQd80pRQoud]Как скачать приложение 1xbet зеркало на андроид 4.4.2 бесплатно русском android 1хбет 2022[/url] [url=https://www.youtube.com/playlist?list=PLEYHnJIeTThw4XoVG2i6bY72O-tJh2lGI]Зеркало 1xbet рабочее на сегодня прямо сейчас мобильная версия 1хбет на май месяц 2022[/url] [url=https://www.youtube.com/playlist?list=PLaHYt0vf74pUKG_BBQr4usrKkMcL2Lva0]1xbet зеркало рабочее на сегодня прямо сейчас мобильная версия скачать айфон в онлайне 1хбет зеркало 2022[/url] [url=https://www.youtube.com/playlist?list=PLFGhmqZCCaybBqMlHJD09-wPZ4Akv5CZ-]1xbet promo code bonus NEW BONUS - OFFICIAL Promo Code - activex2 1xBet bonus code promo code for bonus Superx2029[/url]
https://www.youtube.com/playlist?list=PLrBQniO4U1-XghX_ylsOeH00TPd8OjtxR https://www.youtube.com/playlist?list=PLTQTywpnAAZ–fZNQEp79Yj0u3bc8zFuI https://www.youtube.com/playlist?list=PL6RoMk3kEphaTvY6V4tNCJVSm7EP7SSCw https://www.youtube.com/playlist?list=PLDYPK6IIVo2maDESz0WdXz4Gc2bwjapv4 https://www.youtube.com/playlist?list=PL6Zx4H29Ype159YiKo97oKOu77JDxdreu https://www.youtube.com/playlist?list=PLczAaV_UUCQTwUownUpjCbOrqOC4J-tze https://www.youtube.com/playlist?list=PLEGEICN3tXCa8zqqZ_NimPhk_GG42vchp https://www.youtube.com/playlist?list=PLnOWI5_hHttlgl9qYwipFbvNm9KSfiG2T https://www.youtube.com/playlist?list=PLMUBWbPc6VSCfn5vkzAdbV1bZs57gvHfA
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url] [url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня [/url] https://vk.com/dostup_k_saitu https://vk.com/bet_bonus_perviy_depozit
DorisBot
Салют Приятно найти единомыщленников И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=03zm0jxV1cg https://www.youtube.com/watch?v=zz5KMXa9wRc https://www.youtube.com/watch?v=gPRwGnEDsRY https://www.youtube.com/watch?v=yGH844DRB4E https://www.youtube.com/watch?v=e8JMjCBBCxA https://www.youtube.com/watch?v=Kd-9SAaRDTs https://www.youtube.com/watch?v=i6BW2pQ_vOE https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8
[url=https://www.youtube.com/watch?v=ZHhmI3sjIN4]How to use accumulator promo code in 1xbet kivabe data truncated for column[/url] [url=https://www.youtube.com/watch?v=PrCOCN3b9IA]How does 1xbet promo code work india registration bonus casino 2022 [/url] [url=https://www.youtube.com/watch?v=5eH41BAc9LY]How does 1xbet promo code work india registration bonus casino 2022 [/url] [url=https://www.youtube.com/watch?v=Hqk5sQSyFnE]1xbet promo code 2022 Somali comment parier avec un code promo sur 1xbet[/url] [url=https://www.youtube.com/watch?v=LYnsyGU20qs]1xbet promo code biggest bonus in 2022 for registration account [/url]
[url=https://www.youtube.com/playlist?list=PLN97qBjyP2o8EeayVlBEibGnbpsWZUmVT]Вынос букмекер 1хбет 1xbet зеркало 1xbet - Зеркало 1xBet – Рабочее зеркало 1хбет – вход на 1xBet 2022[/url] [url=https://www.youtube.com/playlist?list=PLyI-Jj1h2qrTWNf3vAK_GjVJ4m4psxAiL]Рабочее зеркало 1xbet скачать бесплатно приложение на андроид на сегодня 2022 регистрация[/url] [url=https://www.youtube.com/playlist?list=PLb33jupKclhQ0XfdMpvQSNY2Uqi_nEpNM]A free bet from 1xbet promo code - How to make promo points free bet in 1xbet.bangla tutorial[/url]
https://www.youtube.com/playlist?list=PLUSskJ5ufx9d2p2S0a0Ua9xYT4pJHEn https://www.youtube.com/playlist?list=PLb-3dY_4SgCzzmjAG9CIO4uZKp22pFNI5 https://www.youtube.com/playlist?list=PL9MIaq_JzVsLMyEDiC9bfgFaOwQXj69AT https://www.youtube.com/playlist?list=PLfEMVWrXpKIO1XPih-1fOelePC-K7hdkK https://www.youtube.com/playlist?list=PL5omy2sjhgJu6z0Y5N-kzlWmlU9NtUN54 https://www.youtube.com/playlist?list=PLW6H-a-airibdiggokqcqWmxktvwEATsq https://www.youtube.com/playlist?list=PLDVbdpOO4MyhkLUuPhSz4XzrjSxTy6B7P https://www.youtube.com/playlist?list=PLC0Ox2GUooS81oCYy3UjU7VGM0fwtzdI- https://www.youtube.com/playlist?list=PLqar3JZGyXqtNTypu4M0KLht_H_Chogmi
[url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня[/url] [url=https://vk.com/bet_bonus_perviy_depozit]1хбет бонус на первый депозит 100% 1xbet условия[/url] [url=https://vk.com/zerkalo__vhod_alternativniy_sait]1xbet зеркало рабочее вход альтернативный сайт[/url] https://vk.com/dostup_k_saitu https://vk.com/bet_bonus_perviy_depozit
DorisBot
Hi хорошая мысль но Так же хочу добавит
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=OGMzex2lcPk https://www.youtube.com/watch?v=XA0GpOcbOSs https://www.youtube.com/watch?v=ZHhmI3sjIN4 https://www.youtube.com/watch?v=PrCOCN3b9IA https://www.youtube.com/watch?v=5eH41BAc9LY https://www.youtube.com/watch?v=Hqk5sQSyFnE https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8
[url=https://www.youtube.com/watch?v=7sTlCqagP90]How to use free bet promo code in 1xbet accumulator - 1xbet no risk bet bangla[/url] [url=https://www.youtube.com/watch?v=8c7sSUXYe_M]How to use free bet promo code in 1xbet accumulator - 1xbet no risk bet bangla[/url] [url=https://www.youtube.com/watch?v=RXnggiHXYO4]1XBET free bet promo code use - no risk bet 1XBET hindi 2022 today[/url] [url=https://www.youtube.com/watch?v=s1dwJtOs9Ek]1xbet promo codes in Pakistan 2022 Hindi Nepal new code promo 1xbet 2022 India[/url]
[url=https://www.youtube.com/playlist?list=PL7XcMMshdq_rxS5Bw4eL3k9tqzRt98sBw]1xbet рабочее зеркало на сегодняшний день актуальное зеркало 1хбет играть сейчас 2022[/url] [url=https://www.youtube.com/playlist?list=PLv341sX3X30qWBS_2w4lVUJts9QGIGPCE]Yow to Create/Make your own promo code in 1xbet/Melbet 2022 [ Hindi Urdu] Official promo code 1xbet. Free 1xbet bonus 2022 1xbet 2022[/url] [url=https://www.youtube.com/playlist?list=PLbHEGmlZUi63dBxl9kntDgEQPEIjouyau]Code promo 1xbet 2022 Maroc - 1xbet promo code 1xbet promo code 2022 1xbet code promo 1xbet BONUS FREE HINDI[/url] [url=https://www.youtube.com/playlist?list=PLXpIHp3B-iXQLcgXCoeW7R9X_pHLKvCxK]Как скачать зеркало 1xbet на андроид казино зеркало мобильная версия один икс бэд май 2022[/url] [url=https://www.youtube.com/playlist?list=PLS1Bff15cYD2fGuCr3gFZVg8rPbKdrCFp]1XBET регистрация по телефону и вход в личный кабинет как зарегистрироваться по номеру телефона - 1xbet регистрация по телефону игровые автоматы[/url]
https://www.youtube.com/playlist?list=PLdX_HbqGQFWSshTXjoVBUUD91sKgkAsip https://www.youtube.com/playlist?list=PLiu2X3QBr87zhKzyQwmB_Cpe9HyP1TjYY https://www.youtube.com/playlist?list=PLqqnMYzfeDg4QD6HmHQ5UgYEUan4anEM8 https://www.youtube.com/playlist?list=PLjAsegiMOATkuJkwXY7tJGAznx3cNuOf4 https://www.youtube.com/playlist?list=PLvibuTWUKNStxbPddi1QGFkQd80pRQoud https://www.youtube.com/playlist?list=PLEYHnJIeTThw4XoVG2i6bY72O-tJh2lGI https://www.youtube.com/playlist?list=PLaHYt0vf74pUKG_BBQr4usrKkMcL2Lva0 https://www.youtube.com/playlist?list=PLFGhmqZCCaybBqMlHJD09-wPZ4Akv5CZ-
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url] [url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня [/url] https://vk.com/dostup_k_saitu https://vk.com/bet_bonus_perviy_depozit
DorisBot
Здрасте хорошая мысль но Так вот
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=d2Lo0oQdU4s https://www.youtube.com/watch?v=uQrqGrcOA68 https://www.youtube.com/watch?v=Cy1SDz7HHq4 https://www.youtube.com/watch?v=gzYfo2U8Bko https://www.youtube.com/watch?v=z68_ZsspLSU https://www.youtube.com/watch?v=xfR0jkYlrKM https://www.youtube.com/watch?v=ld7-fq1JIgM https://www.youtube.com/watch?v=j7Fb6_kkWVc https://www.youtube.com/watch?v=OGMzex2lcPk https://www.youtube.com/watch?v=XA0GpOcbOSs
[url=https://www.youtube.com/watch?v=OGMzex2lcPk]Best promo code for 1xbet bangladesh 2022 free for registration android[/url] [url=https://www.youtube.com/watch?v=XA0GpOcbOSs]How to use accumulator promo code in 1xbet kivabe data truncated for column[/url] [url=https://www.youtube.com/watch?v=ZHhmI3sjIN4]How to use accumulator promo code in 1xbet kivabe data truncated for column[/url] [url=https://www.youtube.com/watch?v=PrCOCN3b9IA]How does 1xbet promo code work india registration bonus casino 2022 [/url] [url=https://www.youtube.com/watch?v=5eH41BAc9LY]How does 1xbet promo code work india registration bonus casino 2022 [/url]
[url=https://www.youtube.com/playlist?list=PLUSskJ5ufx9d2p2S0a0Ua9xYT4pJHEn]1XBET promo code in Pakistan 2022 - 1xbet promo code promo code 1xbet 1xbet promo code 2022[/url] [url=https://www.youtube.com/playlist?list=PLb-3dY_4SgCzzmjAG9CIO4uZKp22pFNI5]Code Promo 1xbet 2022 Senegal, Maroc, Cameroun, Somali, Kenya, Togo Graduation Slogans 2022[/url] [url=https://www.youtube.com/playlist?list=PL9MIaq_JzVsLMyEDiC9bfgFaOwQXj69AT]1XBET регистрация по телефону депозита Украина - как зарегистрироваться в 1xbet на телефоне 2022[/url]
https://www.youtube.com/playlist?list=PLUNnfoTVDWlW1ecYLz_KAYeZFO4s-WYCl https://www.youtube.com/playlist?list=PL8jT0xe-W10AjG4T3AbsJpgeXva_3YwCD https://www.youtube.com/playlist?list=PLEXEd3IuMeL7z5GII87ua6aOYBiGjCDdF https://www.youtube.com/playlist?list=PLM4oyV2WJCEZiLAy3meEdoY2yQJwPfKmN https://www.youtube.com/playlist?list=PL3FnVLICF57sMmRCy_3RVLC_ZxQQzbiXU https://www.youtube.com/playlist?list=PLSjH784sblPXN6_xhvL6IWpkm-5sL9O-n https://www.youtube.com/playlist?list=PLCZ3ZRpd5c10fQIz-447wmirPmJ0hOPwu https://www.youtube.com/playlist?list=PL8lOSh_cY1BXLk7ocEUttl6CHOQ4wAreP https://www.youtube.com/playlist?list=PLG2PnRjcJGQqC41F5U1GzXzHt27Y1uaku
[url=https://vk.com/zerkalo_registraciya_1x]ЗЕРКАЛО 1xbet регистрация на сайте в один клик[/url] [url=https://vk.com/1x_zerkalo_rabotayuschee]Зеркало 1xbet работающее на сегодня прямо сейчас[/url]
https://vk.com/zerkalo_rabochee_1x https://vk.com/dostup_k_saitu
DorisBot
Здрасте со свем согласен и даже более Так же по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=ZHhmI3sjIN4 https://www.youtube.com/watch?v=PrCOCN3b9IA https://www.youtube.com/watch?v=5eH41BAc9LY https://www.youtube.com/watch?v=Hqk5sQSyFnE https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=7sTlCqagP90 https://www.youtube.com/watch?v=8c7sSUXYe_M https://www.youtube.com/watch?v=RXnggiHXYO4 https://www.youtube.com/watch?v=s1dwJtOs9Ek
[url=https://www.youtube.com/watch?v=Kd-9SAaRDTs]How to create own promo code in 1xbet - How to bet with promo code on 1xbet[/url] [url=https://www.youtube.com/watch?v=i6BW2pQ_vOE]How to see our promo code in 1xbet - how to use another promo code in 1xbet[/url] [url=https://www.youtube.com/watch?v=OGMzex2lcPk]Best promo code for 1xbet bangladesh 2022 free for registration android[/url] [url=https://www.youtube.com/watch?v=XA0GpOcbOSs]How to use accumulator promo code in 1xbet kivabe data truncated for column[/url]
[url=https://www.youtube.com/playlist?list=PL7XcMMshdq_rxS5Bw4eL3k9tqzRt98sBw]1xbet рабочее зеркало на сегодняшний день актуальное зеркало 1хбет играть сейчас 2022[/url] [url=https://www.youtube.com/playlist?list=PLv341sX3X30qWBS_2w4lVUJts9QGIGPCE]Yow to Create/Make your own promo code in 1xbet/Melbet 2022 [ Hindi Urdu] Official promo code 1xbet. Free 1xbet bonus 2022 1xbet 2022[/url] [url=https://www.youtube.com/playlist?list=PLbHEGmlZUi63dBxl9kntDgEQPEIjouyau]Code promo 1xbet 2022 Maroc - 1xbet promo code 1xbet promo code 2022 1xbet code promo 1xbet BONUS FREE HINDI[/url] [url=https://www.youtube.com/playlist?list=PLXpIHp3B-iXQLcgXCoeW7R9X_pHLKvCxK]Как скачать зеркало 1xbet на андроид казино зеркало мобильная версия один икс бэд май 2022[/url] [url=https://www.youtube.com/playlist?list=PLS1Bff15cYD2fGuCr3gFZVg8rPbKdrCFp]1XBET регистрация по телефону и вход в личный кабинет как зарегистрироваться по номеру телефона - 1xbet регистрация по телефону игровые автоматы[/url]
https://www.youtube.com/playlist?list=PLN97qBjyP2o8EeayVlBEibGnbpsWZUmVT https://www.youtube.com/playlist?list=PLyI-Jj1h2qrTWNf3vAK_GjVJ4m4psxAiL https://www.youtube.com/playlist?list=PLb33jupKclhQ0XfdMpvQSNY2Uqi_nEpNM https://www.youtube.com/playlist?list=PLTzh1NeGvnokHAOHCGxIXoS9AC8j0VQiw https://www.youtube.com/playlist?list=PL4JzUa3785d2VK_1qO3wX2WO7rpCI8IyJ https://www.youtube.com/playlist?list=PLhk4BTkxV4-qt_KwbjU8zwep-RVtvakug https://www.youtube.com/playlist?list=PLKBVJtK4p7XhKNT1LNQ9hE7Nt_cBs_sXs https://www.youtube.com/playlist?list=PLmKYae9PeQwAlFV7DARRZUSV8mnOB79UA
[url=https://vk.com/zerkalo_registraciya_1x]ЗЕРКАЛО 1xbet регистрация на сайте в один клик[/url] [url=https://vk.com/1x_zerkalo_rabotayuschee]Зеркало 1xbet работающее на сегодня прямо сейчас[/url]
https://vk.com/zerkalo__vhod_alternativniy_sait https://vk.com/zerkalo_registraciya_1x https://vk.com/1x_zerkalo_rabotayuschee
DorisBot
Привет согласен и даже более Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=OGMzex2lcPk https://www.youtube.com/watch?v=XA0GpOcbOSs https://www.youtube.com/watch?v=ZHhmI3sjIN4 https://www.youtube.com/watch?v=PrCOCN3b9IA https://www.youtube.com/watch?v=5eH41BAc9LY https://www.youtube.com/watch?v=Hqk5sQSyFnE https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8
[url=https://www.youtube.com/watch?v=d2Lo0oQdU4s]How to see our promo code in 1xbet - how to use another promo code in 1xbet[/url] [url=https://www.youtube.com/watch?v=uQrqGrcOA68]How to get promo code in 1xbet telugu - how to generate 1xbet promo code[/url] [url=https://www.youtube.com/watch?v=Cy1SDz7HHq4]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url] [url=https://www.youtube.com/watch?v=gzYfo2U8Bko]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url] [url=https://www.youtube.com/watch?v=z68_ZsspLSU]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url]
[url=https://www.youtube.com/playlist?list=PL6Zx4H29Ype159YiKo97oKOu77JDxdreu]Как зайти на 1xbet через зеркало сегодня войти через зеркало вк прямо сейчас на сайт 1хбет[/url] [url=https://www.youtube.com/playlist?list=PLczAaV_UUCQTwUownUpjCbOrqOC4J-tze]Code promo 1xbet 2022 togo - code promo 1xbet 2022 Kenya[/url] [url=https://www.youtube.com/playlist?list=PLEGEICN3tXCa8zqqZ_NimPhk_GG42vchp]1xBet официальный сайт регистрация Как зарегистрироваться в БК 1xBet с телефона - 1xbet регистрация по телефону мобильная версия[/url] [url=https://www.youtube.com/playlist?list=PLnOWI5_hHttlgl9qYwipFbvNm9KSfiG2T]1XBET promo code Nepal 2022 - 1xbet promo codes 2022 new[/url] [url=https://www.youtube.com/playlist?list=PLMUBWbPc6VSCfn5vkzAdbV1bZs57gvHfA]1XBET promo code 2022 Somali - 1xbet promo code 1xbet promo code for registration 1xbet promo code 2022[/url]
https://www.youtube.com/playlist?list=PLUNnfoTVDWlW1ecYLz_KAYeZFO4s-WYCl https://www.youtube.com/playlist?list=PL8jT0xe-W10AjG4T3AbsJpgeXva_3YwCD https://www.youtube.com/playlist?list=PLEXEd3IuMeL7z5GII87ua6aOYBiGjCDdF https://www.youtube.com/playlist?list=PLM4oyV2WJCEZiLAy3meEdoY2yQJwPfKmN https://www.youtube.com/playlist?list=PL3FnVLICF57sMmRCy_3RVLC_ZxQQzbiXU https://www.youtube.com/playlist?list=PLSjH784sblPXN6_xhvL6IWpkm-5sL9O-n https://www.youtube.com/playlist?list=PLCZ3ZRpd5c10fQIz-447wmirPmJ0hOPwu https://www.youtube.com/playlist?list=PL8lOSh_cY1BXLk7ocEUttl6CHOQ4wAreP https://www.youtube.com/playlist?list=PLG2PnRjcJGQqC41F5U1GzXzHt27Y1uaku
[url=https://vk.com/zerkalo_rabochee_1x]1xbet зеркало рабочее сегодня прямо сейчас[/url] [url=https://vk.com/dostup_k_saitu]Доступ сайту 1xbet зеркало рабочее сегодня [/url] https://vk.com/dostup_k_saitu https://vk.com/bet_bonus_perviy_depozit
Danuta
If you desire to grow your experience just keep visiting this web site and be updated with the most recent gossip posted here.
Roseanna
Hey there! I realize this is sort of off-topic however I had to ask. Does operating a well-established blog like yours require a massive amount work? I’m brand new to writing a blog however I do write in my journal every day. I’d like to start a blog so I can easily share my personal experience and views online. Please let me know if you have any suggestions or tips for brand new aspiring blog owners. Thankyou!
Ada
There is definately a lot to learn about this issue. I like all the points you’ve made.
Garry
Excellent post. I was checking continuously this blog and I am impressed! Very useful info specifically the last part : ) I care for such info a lot. I was looking for this particular info for a long time. Thank you and best of luck.
Rickey
Холтер ЭКГ
DorisBot
Салют со свем согласен и даже более И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=OGMzex2lcPk https://www.youtube.com/watch?v=XA0GpOcbOSs https://www.youtube.com/watch?v=ZHhmI3sjIN4 https://www.youtube.com/watch?v=PrCOCN3b9IA https://www.youtube.com/watch?v=5eH41BAc9LY https://www.youtube.com/watch?v=Hqk5sQSyFnE https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8
[url=https://www.youtube.com/watch?v=d2Lo0oQdU4s]How to see our promo code in 1xbet - how to use another promo code in 1xbet[/url] [url=https://www.youtube.com/watch?v=uQrqGrcOA68]How to get promo code in 1xbet telugu - how to generate 1xbet promo code[/url] [url=https://www.youtube.com/watch?v=Cy1SDz7HHq4]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url] [url=https://www.youtube.com/watch?v=gzYfo2U8Bko]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url] [url=https://www.youtube.com/watch?v=z68_ZsspLSU]Best promo code for 1xbet Bangladesh 2022 free for registration Android[/url]
[url=https://www.youtube.com/playlist?list=PLHi4_ZIOpSWclC944IwxL_olsaueHxBOv]Как скачать зеркало 1xbet на андроид казино зеркало мобильная версия один икс бэд сентябрь октябрь ноябрь декабрь 2022[/url] [url=https://www.youtube.com/playlist?list=PL6uJP2LK7IrbLzbCYCKWXPzSwkXsK20VS]1xbet promo code 2022 Bangla - 1xbet promo code promo code 1xbet 1xbet promo code 2022[/url] [url=https://www.youtube.com/playlist?list=PLtWkM0udMxKWIKOub-Wr9hopGDk1Pwd7B]1XBET регистрацию по телефону депозита Украина - как зарегистрироваться в 1xbet на телефоне 2022[/url] [url=https://www.youtube.com/playlist?list=PLzQ27XYItO-a_LmprGpDfxSuymNpYUume]XBET promo code India 2022 - code promo 1xbet 2022 English[/url] [url=https://www.youtube.com/playlist?list=PLBNRuEFg2nVS9FCISN2oUGVVicpSIbc_J]Code promo 1XBET 2022 cote d’ivoire - 1xBet promo code 2022 BIG BONUS OFFICIAL Promo Code 1xbet 425[/url]
https://www.youtube.com/playlist?list=PLUSskJ5ufx9d2p2S0a0Ua9xYT4pJHEn https://www.youtube.com/playlist?list=PLb-3dY_4SgCzzmjAG9CIO4uZKp22pFNI5 https://www.youtube.com/playlist?list=PL9MIaq_JzVsLMyEDiC9bfgFaOwQXj69AT https://www.youtube.com/playlist?list=PLfEMVWrXpKIO1XPih-1fOelePC-K7hdkK https://www.youtube.com/playlist?list=PL5omy2sjhgJu6z0Y5N-kzlWmlU9NtUN54 https://www.youtube.com/playlist?list=PLW6H-a-airibdiggokqcqWmxktvwEATsq https://www.youtube.com/playlist?list=PLDVbdpOO4MyhkLUuPhSz4XzrjSxTy6B7P https://www.youtube.com/playlist?list=PLC0Ox2GUooS81oCYy3UjU7VGM0fwtzdI- https://www.youtube.com/playlist?list=PLqar3JZGyXqtNTypu4M0KLht_H_Chogmi
[url=https://vk.com/zerkalo_registraciya_1x]ЗЕРКАЛО 1xbet регистрация на сайте в один клик[/url] [url=https://vk.com/1x_zerkalo_rabotayuschee]Зеркало 1xbet работающее на сегодня прямо сейчас[/url]
https://vk.com/zerkalo__vhod_alternativniy_sait https://vk.com/zerkalo_registraciya_1x https://vk.com/1x_zerkalo_rabotayuschee
DorisBot
Здравствуйте хорошая мысль но И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=03zm0jxV1cg https://www.youtube.com/watch?v=zz5KMXa9wRc https://www.youtube.com/watch?v=gPRwGnEDsRY https://www.youtube.com/watch?v=yGH844DRB4E https://www.youtube.com/watch?v=e8JMjCBBCxA https://www.youtube.com/watch?v=Kd-9SAaRDTs https://www.youtube.com/watch?v=i6BW2pQ_vOE https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8
[url=https://www.youtube.com/watch?v=ZHhmI3sjIN4]How to use accumulator promo code in 1xbet kivabe data truncated for column[/url] [url=https://www.youtube.com/watch?v=PrCOCN3b9IA]How does 1xbet promo code work india registration bonus casino 2022 [/url] [url=https://www.youtube.com/watch?v=5eH41BAc9LY]How does 1xbet promo code work india registration bonus casino 2022 [/url] [url=https://www.youtube.com/watch?v=Hqk5sQSyFnE]1xbet promo code 2022 Somali comment parier avec un code promo sur 1xbet[/url] [url=https://www.youtube.com/watch?v=LYnsyGU20qs]1xbet promo code biggest bonus in 2022 for registration account [/url]
[url=https://www.youtube.com/playlist?list=PLTzh1NeGvnokHAOHCGxIXoS9AC8j0VQiw]1xbet promo code bonus. Get free bonus 1xbet 1xbet bonus code OFFICIAL Biggest Bonus 2022 1xbet promo code Working 100%[/url] [url=https://www.youtube.com/playlist?list=PL4JzUa3785d2VK_1qO3wX2WO7rpCI8IyJ]1XBET promo code for bonus Use XLIKE22 for registration 1xBET PROMO CODE 1XBET BONUS 1xbet FREE official promo code 2022 year[/url] [url=https://www.youtube.com/playlist?list=PLhk4BTkxV4-qt_KwbjU8zwep-RVtvakug]1xbet promo code bonus NEW BONUS - OFFICIAL Promo Code - activex2 1xBet bonus code promo code for bonus Superx2029[/url] [url=https://www.youtube.com/playlist?list=PLKBVJtK4p7XhKNT1LNQ9hE7Nt_cBs_sXs]Как скачать приложение 1xbet зеркало на андроид 4.4.2 бесплатно русском android 1хбет 2022[/url] [url=https://www.youtube.com/playlist?list=PLmKYae9PeQwAlFV7DARRZUSV8mnOB79UA]Как скачать 1xbet на Андроид последняя версия зеркало работающее официальный сайт казино зеркало 1хбет 2022[/url]
https://www.youtube.com/playlist?list=PLZHO1qxSiWyaJG6_YCWCyxCjGluSmvv6B https://www.youtube.com/playlist?list=PLn4WkOv9zpeXQsspQSEwPudnY13Ghe2RX https://www.youtube.com/playlist?list=PLHnBmYYtiE6HFxtI81eyPy_HBh08A2K9N https://www.youtube.com/playlist?list=PLrwoWoOGd4wT8-uGifJ3p0LInXmI6hXfn
[url=https://vk.com/zerkalo_registraciya_1x]ЗЕРКАЛО 1xbet регистрация на сайте в один клик[/url] [url=https://vk.com/1x_zerkalo_rabotayuschee]Зеркало 1xbet работающее на сегодня прямо сейчас[/url]
https://vk.com/dostup_k_saitu https://vk.com/bet_bonus_perviy_depozit
Hudson
Link exchange is nothing else however it is simply placing the other person’s weblog link on your page at suitable place and other person will also do same in support of you.
Penelope
I like the valuable information you provide in your articles. I will bookmark your weblog and check again here regularly.
I am quite certain I’ll learn a lot of new stuff right here! Best of luck for the next!
Danny
You also acknowledge that your agrement to get these messages is not needed as a condition to acquire goods or services.
KendraEmurf
Салют хорошая мысль но Вот что я хочу добавить
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=LYnsyGU20qs https://www.youtube.com/watch?v=PQu5z8kzS90 https://www.youtube.com/watch?v=rtX865F7jAo https://www.youtube.com/watch?v=zi_rpdpjLo8 https://www.youtube.com/shorts/UQR6a9gnt1s
https://www.youtube.com/playlist?list=PLRcnUiG5ZvBAkZ1kLwOgJvlkbOpgayKLW https://www.youtube.com/playlist?list=PLRcnUiG5ZvBD0pQAF3M55iGZwGZvETiQG https://www.youtube.com/playlist?list=PLRcnUiG5ZvBBxxLF0L0GCcSb6LulGFewU https://www.youtube.com/playlist?list=PLRcnUiG5ZvBAiBMOYAMECWuTPuoBXM0Uv https://www.youtube.com/playlist?list=PLRcnUiG5ZvBD0Q1ao7kkblJlI2vCkNjuB https://www.youtube.com/playlist?list=PLRcnUiG5ZvBAsIXBJTe8PAUe58zJt3M-G https://www.youtube.com/playlist?list=PLRcnUiG5ZvBDD5R_6AY2SSIcIe2J3bVdx https://www.youtube.com/playlist?list=PLRcnUiG5ZvBCacBJurDWIUErdX89bvBt1 https://www.youtube.com/playlist?list=PLRcnUiG5ZvBDFKQv0iEYdJBixD9iqUcRh
https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBJiG7pECb7QDPwpDPhJlfCh https://www.youtube.com/playlist?list=PLq4BI-2vvhBJORFMgQteH3OGrZFAQRruY https://www.youtube.com/playlist?list=PLq4BI-2vvhBItDs8megR3fmRAnpDhPLTc https://www.youtube.com/playlist?list=PLq4BI-2vvhBKhGxV7sq0r8O2PJAWE4VH7 https://www.youtube.com/playlist?list=PLq4BI-2vvhBI-Ab6axauG7f0PMciZvhxo https://www.youtube.com/playlist?list=PLq4BI-2vvhBLxpjDLTpoO1htzns24L7XQ https://www.youtube.com/playlist?list=PLq4BI-2vvhBJfSYQYyuDb9Jy7Bdz155kS https://www.youtube.com/playlist?list=PLq4BI-2vvhBIwDotoD1zozfoxAT1vioi3
Romeo
Hello great blog! Does running a blog like this take a lot of work?
I have absolutely no knowledge of coding but I was hoping to start my own blog in the near future. Anyhow, if you have any ideas or tips for new blog owners please share. I understand this is off topic however I simply had to ask. Kudos!
JoanneHek
Здравствуйте со свем согласен И вот это тоже по теме
РАБОЧИЕ ЗЕРКАЛА НА 1ХBET http://bit.ly/37SxqIa - смотрите вкладку АКЦИИ
https://www.youtube.com/watch?v=ZGiZvXquPcQ https://www.youtube.com/watch?v=g_d0Zyl2VqE https://www.youtube.com/watch?v=vIZD9oOl0tg https://www.youtube.com/watch?v=HLrq-tjYTx8 https://www.youtube.com/watch?v=TMBWt3Xf5xw https://www.youtube.com/watch?v=fieWXlMi01o https://www.youtube.com/watch?v=GhBNvgzg5oc
[url=https://www.youtube.com/watch?v=LYnsyGU20qs]1xbet promo code biggest bonus in 2022 for registration account [/url] [url=https://www.youtube.com/watch?v=PQu5z8kzS90]1xbet promo code biggest bonus in 2022 for registration account [/url] [url=https://www.youtube.com/watch?v=rtX865F7jAo]1xbet me promo code kaise dale - comment creer son code promo 1xbet[/url] [url=https://www.youtube.com/watch?v=zi_rpdpjLo8]1xbet me promo code kaise dale - comment creer son code promo 1xbet [/url] [url=https://www.youtube.com/shorts/UQR6a9gnt1s]Как скачать 1хбет зеркало на айфон бесплатно 1xbet на андроид на русском зеркало 2022[/url]
1xbet loss cover tricks 1xbet prediction Bangla Tutorial online tips Bangla[/url] [url=https://www.youtube.com/playlist?list=PLmsxJP-MOi-s-MRk2iwog9A_XWkJ2Vdfq]1xBet promo code Bangladesh 1xbet Bangla Tutorial - 1XBET bangla tutorial promo code[/url] [url=https://www.youtube.com/playlist?list=PLAzjLuk66MHBgXWABVLB8WCoNA8Cf19ze]1xbet.how to open 1xbet account bangla.1xbet bangla tutorial.how to create 1xbet account 1xbet signup[/url]
https://www.youtube.com/playlist?list=PLEBUODUctZO9eTz8LNHQLeYsTcXfpaA1B https://www.youtube.com/playlist?list=PLT2jWTYv4X1auK8zsKdsa0xlxtV05zQt4 https://www.youtube.com/playlist?list=PLrjJ6mR6zrZryGb8UM4uskhdl9qeZE414 https://www.youtube.com/playlist?list=PLmjEqNxuviBSwC72aSIy4yOQzdFk0HekL https://www.youtube.com/playlist?list=PLooJc4ieNOfA2J8YTtRNEOQ3qh80TuK0I https://www.youtube.com/playlist?list=PLxBzrFl7ByegkbpykES7tjWLXyYAj2Ah6 https://www.youtube.com/playlist?list=PLvF8VgIhx5ATFIDTi3ThGtV2diSPrHOKb https://www.youtube.com/playlist?list=PLGPmFPQotcMekAnyUUusNupoNAjk0Rt0T
Sima
Just desire to say your article is as amazing. The clarity for your submit is simply cool and that i could think you are a professional on this subject.
Well along with your permission let me to take hold of your RSS feed to stay up to date with imminent post. Thanks a million and please continue the enjoyable work.
Addie
My spouse and I stumbled over here from a different website and thought I should check things out. I like what I see so i am just following you. Look forward to exploring your web page repeatedly.
Garfield
Just want to say your article is as astounding.
The clarity to your publish is just spectacular and that i can suppose you are an expert on this subject. Fine together with your permission let me to grab your RSS feed to stay up to date with approaching post. Thank you a million and please continue the enjoyable work.
Nicholas
Your method of telling the whole thing in this piece of writing is in fact fastidious, all be able to effortlessly be aware of it, Thanks a lot.
Adrienne
Appreciate this post. Let me try it out.
Nicole
I’m truly enjoying the design and layout of your website. It’s a very easy on the eyes which makes it much more pleasant for me to come here and visit more often. Did you hire out a developer to create your theme? Great work!
Zandra
Hello to every body, it’s my first visit of this web site; this blog includes remarkable and actually excellent data in favor of readers.
Edith
Excellent write-up. I absolutely appreciate this site. Continue the good work!
Oliva
Yes! Finally something about car finance calculator.
Kathy
Its like you read my thoughts! You appear to understand a lot approximately this, such as you wrote the book in it or something. I feel that you just can do with a few p.c. to drive the message home a little bit, but other than that, this is wonderful blog. An excellent read. I’ll definitely be back.
Laura
You have made some really good points there. I checked on the web to learn more about the issue and found most people will go along with your views on this site.
Sarah
Heya! I know this is kind of off-topic but I had to ask. Does operating a well-established website such as yours require a large amount of work? I’m brand new to operating a blog but I do write in my journal daily. I’d like to start a blog so I will be able to share my personal experience and thoughts online. Please let me know if you have any recommendations or tips for brand new aspiring bloggers. Thankyou!
Veda
Appreciation to my father who stated to me on the topic of this blog, this website is really remarkable.
Sammy
An impressive share! I have just forwarded this onto a colleague who had been conducting a little research on this. And he in fact bought me breakfast due to the fact that I found it for him… lol. So let me reword this…. Thank YOU for the meal!! But yeah, thanx for spending some time to talk about this subject here on your site.
Thomassok
Hi there! [url=https://onlinesxpharmacy.quest/#/]vicodin online pharmacy[/url] beneficial site https://pharmacyonlinexp.quest/#
Maik
Having read this I believed it was really enlightening. I appreciate you taking the time and effort to put this article together. I once again find myself spending a significant amount of time both reading and leaving comments. But so what, it was still worth it!
Leave a comment
Your email address will not be published. Required fields are marked *