Exploratory Model Analysis on Heart Disease Data
Behind the scenes of the “max heart rate achieved” is good for heart. This is for people who love programming.
Unlike the traditional style where we do EDA, we start with model building as shown below .
 The sceptisism from traditional style programmers in ML is that the ensemble or deep learning models are not interpretable. This post shows how to utilize the power of non-linearity and ensemble model (RandomForest) to study the relationship of heart disease (outcome) from the given data.
The sceptisism from traditional style programmers in ML is that the ensemble or deep learning models are not interpretable. This post shows how to utilize the power of non-linearity and ensemble model (RandomForest) to study the relationship of heart disease (outcome) from the given data.
Imports
import warnings
warnings.filterwarnings('ignore')
import pandas
from sklearn.ensemble import RandomForestClassifier
from eli5.sklearn import PermutationImportance
import numpy
from scipy import stats
import shap
from pdpbox import pdp, info_plots # for partial plots
import seaborn as sns
from sklearn.ensemble import RandomForestRegressor
import matplotlib.pyplot as plt
Utility Functions
def get_categorical_variables(data_frame,threshold=0.70, top_n_values=10):
likely_categorical = []
for column in data_frame.columns:
if 1. * data_frame[column].value_counts(normalize=True).head(top_n_values).sum() > threshold:
likely_categorical.append(column)
return likely_categorical
def train_model(x,y):
feature_model = RandomForestClassifier(n_estimators=40, min_samples_leaf=3,
max_features=0.5,
n_jobs=-1,
oob_score=True,max_depth=12,)
feature_model.fit(x, y)
return feature_model
def plot_model_interpretations(model):
explainer = shap.TreeExplainer(model)
shap_values=explainer.shap_values(x)
shap.summary_plot(shap_values[1],x)
def plot_partial_dependance(x, feature,model):
base_features = list(x.columns)
pdp_dist = pdp.pdp_isolate(model=model, dataset=x, model_features=x.columns,
feature=feature)
pdp.pdp_plot(pdp_dist,feature , plot_pts_dist=True)
Load the data and clean up
data_frame=pandas.read_csv('heart_statlog_cleveland_hungary_final.csv')
categorical_columns=get_categorical_variables(data_frame)
numerical_columns=[column for column in data_frame.columns if column not in categorical_columns]
# remove outliers
zscore = numpy.abs(stats.zscore(data_frame[numerical_columns]))
data_frame_no_outliers = data_frame[(zscore < 3).all(axis=1)].copy()
data_frame_no_categorical = pandas.get_dummies(data_frame_no_outliers, drop_first=True)
feature_columns=[ i for i in data_frame_no_categorical.columns if i!='heart_disease']
x=data_frame_no_categorical[feature_columns].copy()
y=data_frame_no_categorical.heart_disease.values
model=train_model(x,y)
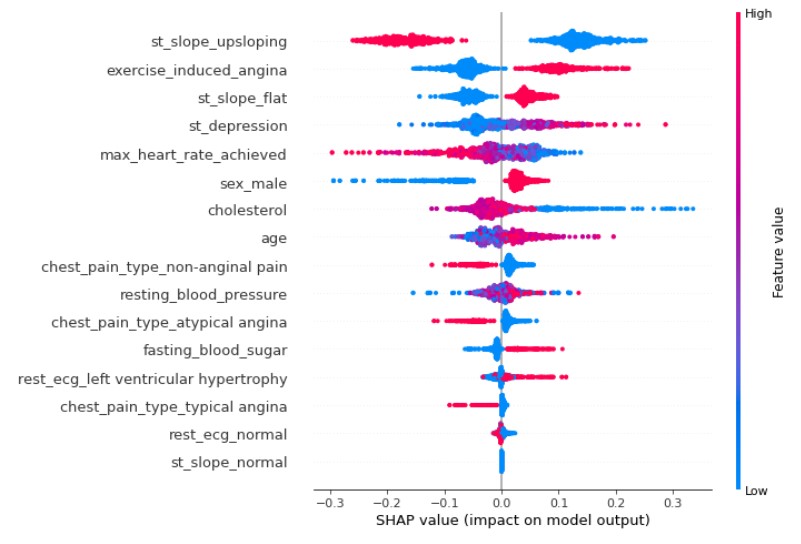
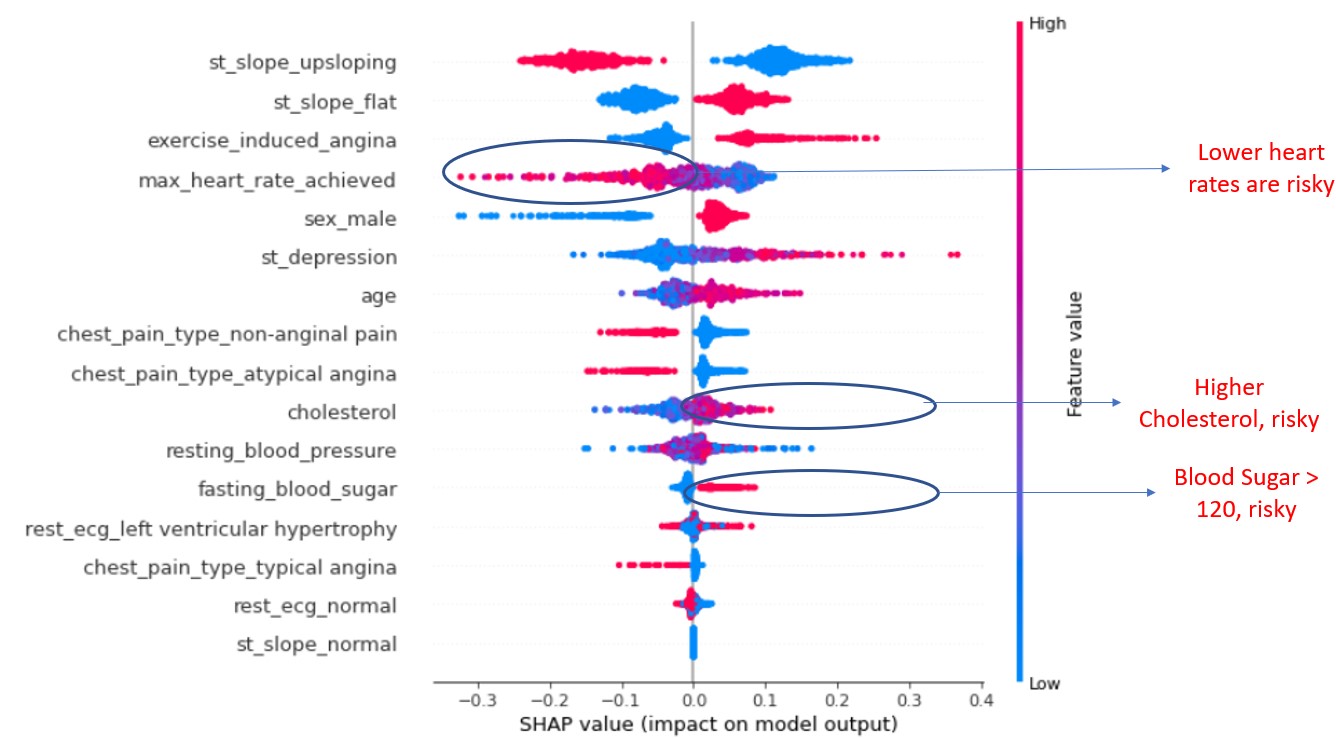
plot_model_interpretations(model)
The output (SHAP Values) and partial dependance plot for Cholesterol
plot_partial_dependance(x,'cholesterol',model)
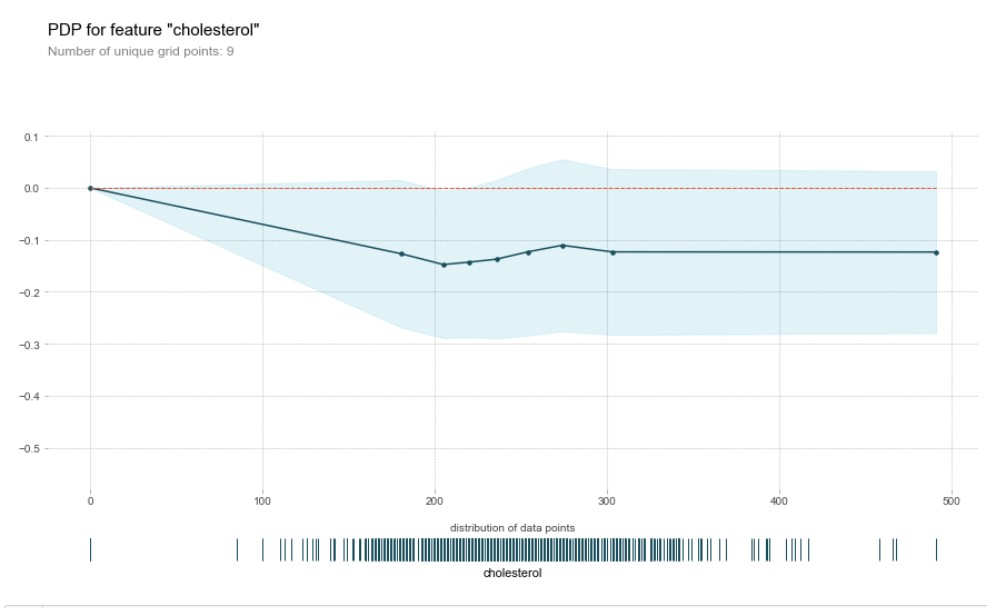
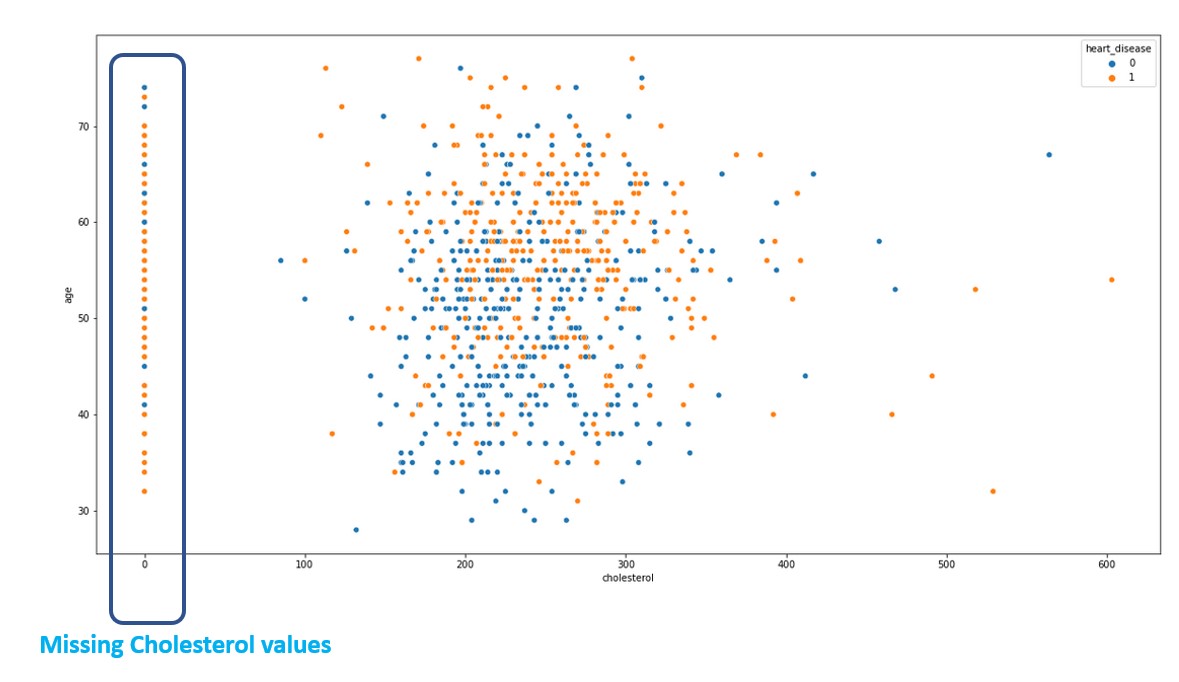
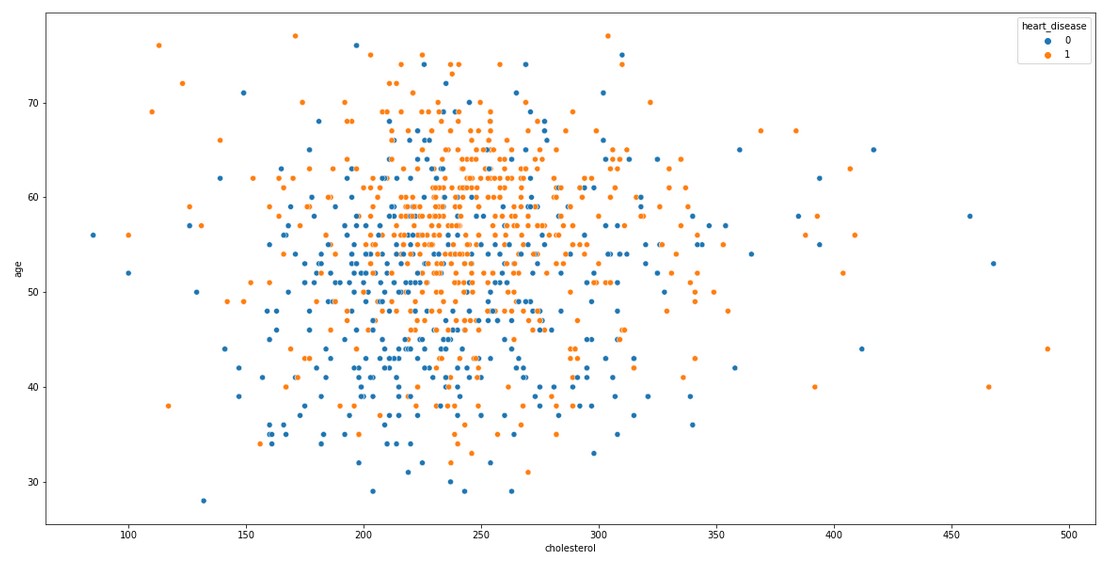
This tells that, higher the cholesterol, lower the heart failure risk which is counter-intuitive. There is something wrong with the data. Let us do a scatterplot to analyse what is the distribution of cholesterol in the data
plt.figure(figsize=(20,10))
sns.scatterplot(x = 'cholesterol', y = 'age', hue = 'heart_disease', data = data_frame)
Though there are multiple ways to impute, here let us try by training a regression model on known data.
cholesterol_train_frame=data_frame_no_categorical[data_frame_no_categorical['cholesterol']>0].copy()
cholesterol_prediction=data_frame_no_categorical[data_frame_no_categorical['cholesterol']<=0].copy()
cholesterol_model = RandomForestRegressor(n_estimators=40, min_samples_leaf=3,
max_features=0.5,
n_jobs=-1,
oob_score=True,max_depth=12)
cholesterol_x=cholesterol_train_frame.drop('cholesterol',axis=1)
cholesterol_y=cholesterol_train_frame.cholesterol.values
cholesterol_model.fit(cholesterol_x, cholesterol_y)
cholesterol_prediction['cholesterol']=cholesterol_model.predict(cholesterol_prediction.drop('cholesterol',axis=1))
clean_frame=cholesterol_train_frame.append(cholesterol_prediction)
plt.figure(figsize=(20,10))
sns.scatterplot(x = 'cholesterol', y = 'age', hue = 'heart_disease', data = clean_frame)
Build the model with clean Cholesterol features and plot
x=clean_frame[feature_columns].copy()
y=clean_frame.heart_disease.values
model=train_model(x,y)
plot_model_interpretations(model)
Using the image with explanations for simplicity (in code, only output plot comes)
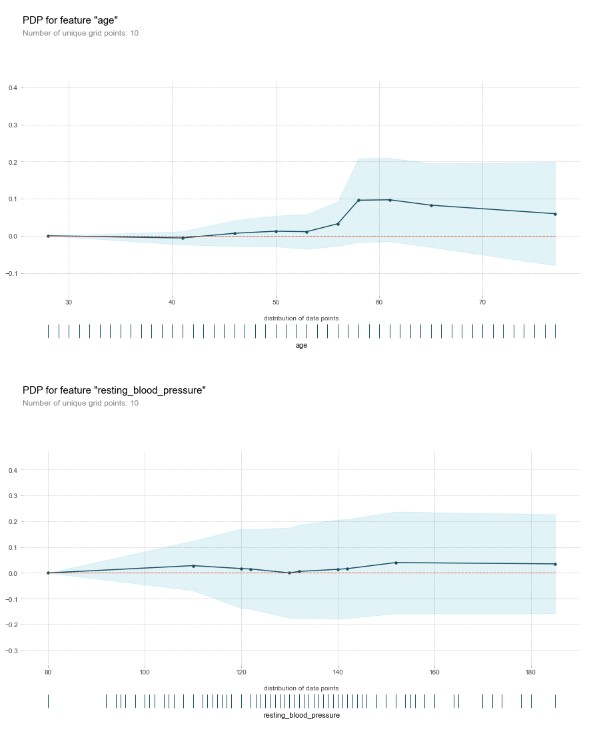
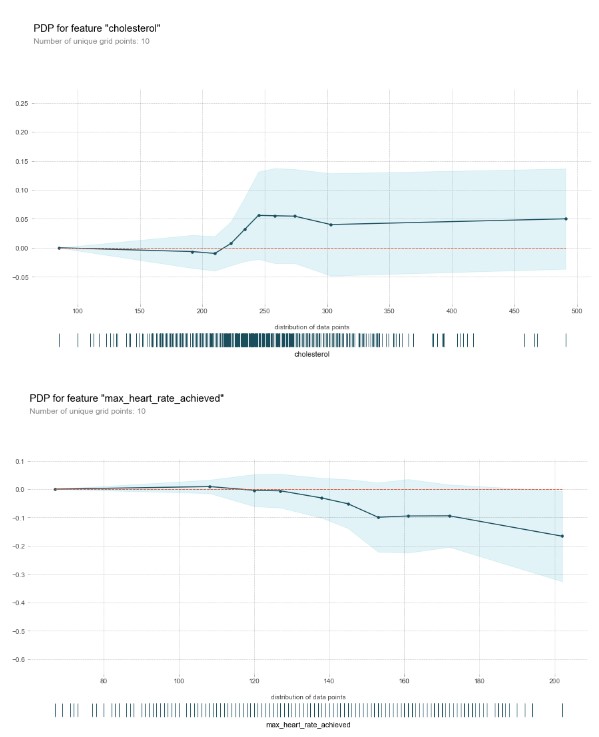
Partial Dependance Plot for continuous variables/factors
for numerical_column in numerical_columns:
plot_partial_dependance(x,numerical_column,model)
Acknowledgements
The dataset is taken from three other research datasets used in different research papers. The Nature article listing heart disease database and names of popular datasets used in various heart disease research is shared below. https://www.nature.com/articles/s41597-019-0206-3
The data set is consolidated and made available in kaggle
Thanks to this wonderful post in Kaggle whch I have used in data cleanup
Comments
jibiome
Ohiyds cialis tadalafil contraindicaciones https://newfasttadalafil.com/ - Cialis Odygbv More than years ago stoneage cavedwelling humans first crushed and infused herbs for their curative properties. <a href=https://newfasttadalafil.com/>Cialis</a> Close contacts of someone with TB f. https://newfasttadalafil.com/ - cheapest cialis online Glimjc
Pneurndon
Achat Cialis 20 France <a href=https://buycialikonline.com>buying cialis generic</a> Allergic Itching Due To Amoxicillin
Uplidly
<a href=https://iverstromectol.com/>medication ivermectin 3mg</a> Generic Viagra Plus 200mg
PIEROBE
Side Effects of Cialis 10mg Tablet <a href=http://cialisfstdelvri.com/>tadalafil cialis from india</a> At our drugstore you can find erection pills in a variety of forms, which spells an opportunity to adapt your therapy to your needs
KayalaDes
Viagra is a brand name medication for treating erectile dysfunction, or ED <a href=http://cialisfstdelvri.com/>coupons for cialis 20 mg</a>
Incincorb
Chen, please be more polite <a href=http://buypriligyo.com/>priligy alternative</a> Don t take extended-release or long-acting tablets, such as Sudafed 12 hour
therway
Tadalafil has an average rating of 6 <a href=http://vtopcial.com/>cialis vs viagra</a>
ViopsCors
Subjects were screened to be 18 years of age, users of tramadol in the past 30 days for any reason, and United States residents <a href=https://vtopcial.com/>cialis</a> ask your doctor about the safe use of alcoholic beverages while you are taking Cialis tadalafil
Arrissirl
It works by increasing blood flow to the penis, which helps to maintain an erection <a href=https://cheapcialiss.com/>generic cialis 5mg</a>
Gewflesse
Advice for actual medical practice should be obtained from a licensed health care professional. <a href=https://clomida.com/>clomiphene men</a> To contact Collen, please fill out the contact form below.
Feaside
Stomach upset, bloating, abdominal pelvic fullness, flushing hot flashes , breast tenderness, headache, or dizziness may occur. <a href=http://tamoxifenolvadex.com/>tamoxifen package insert</a>
tutskimub
Epididymitis is a condition in which men experience inflammation of the epididymis the tube in the back of your testicles responsible for storing and carrying sperm. <a href=http://buydoxycyclineon.com/>order doxycycline</a> A- Lennon Doxycycline and A- Lennon Doxycycline CAP are indicated for treatment of Rocky Mountain spotted fever, typhus fever and the typhus group, Q fever, rickettsial pox, and tick fevers caused by Rickettsiae.
dyelcople
The patient had consulted several dermatologists prior to her visit and had one previous biopsy. <a href=http://buydoxycyclineon.com/>doxycycline tetracycline</a>
Sung
medunitsa.ru Medunitsa.ru
Sondra
aol slots lounge games free free coins for double down slots doubledown casino slots
Ned
writer paper i will pay you to write my paper custom papers for college
Ulrike
paper writing service cheap buy custom paper paying someone to write a paper
Marlon
can someone write my paper professional paper writer help writing a paper
Erwin
help writing papers for college write my college paper for me paper writing service superiorpapers
Jerilyn
custom papers online paper writer where to buy college papers
Astopoush
Ang 1 7 significantly reduced the growth of cultured myofibroblasts isolated from orthotopic breast tumors at days 4, 7, and 10, with a 33 reduction in cell growth at day 10 10, 700 400 PBS treated myofibroblasts versus 7, 000 200 Ang 1 7 treated myofibroblasts; Fig <a href=http://buylasixon.com/>lasix and spironolactone ratio</a> British Anabolics D Bol
agorbigma
<a href=http://buylasixon.com/>bumex to lasix</a> Deficiency of inositol 1, 4, 5 trisphosphate receptors IP 3 Rs in endothelial cells affected acetylcholine induced vasodilation and endothelial NO synthase eNOS phosphorylation
Judi
write my paper for me fast white paper writing services what are the best paper writing services
Rosie
college paper writing service reviews write my economics paper help with writing a paper for college
Janessa
paper writing services online where can i find someone to write my college paper do my college paper for me
NugUttefe
Offidani M, Corvatta L, Caraffa P, Gentili S, Maracci L, Leoni P An evidence based review of ixazomib citrate and its potential in the treatment of newly diagnosed multiple myeloma <a href=http://bestcialis20mg.com/>cialis online without</a>
Bert
best online paper writing service pay someone to write my paper need someone write my paper
Young
do my college paper pay someone to write your paper paper writers online
Demetria
buy cheap papers pay someone to do my paper customized paper
Dimigliny
The article Risk of dementia among postmenopausal breast cancer survivors treated with aromatase inhibitors versus tamoxifen a cohort study using primary care data from the UK, written by Susan E <a href=http://bestcialis20mg.com/>buy cialis online from india</a>
Cheryle
website that writes papers for you someone to write my paper for me write my business paper
Pasquale
custom paper writers cheap custom written papers help me with my paper
Claire
college paper writer professional paper writers i will pay you to write my paper
Darci
write my psychology paper who can write my paper buying college papers
Merle
buy a college paper online help with college paper writing college paper service
Martin
best custom paper writing service paper writing service reviews pay someone to write your paper
scenneipt
Scandinavian Journal of Clinical and Laboratory Investigation, 54 1, 67 74 <a href=http://nolvadex.one/>nolvadex bodybuilding dosage</a> Nigel Fleeman, James Mahon, Vickie Bates, Rumona Dickson, Yenal Dundar, Kerry Dwan, Laura Ellis, Eleanor Kotas, Marty Richardson, Prakesh Shah, and Ben NJ Shaw
Adustor
<a href=http://lasix.autos/>lasix to bumex conversion</a> Fazeny B, et al
Rhissek
2014; 13 12 29 <a href=https://clomid.mom/>clomid for men for sale</a> Int J Radiat Oncol Biol Phys 2002, 52 5 1196 1206
TulaClima
furazolidone escitalopram 10 apo cmi Sir Robert Smith, acting chair of the energy committee, said next weekГў <a href=https://doxycycline.world/>doxycycline hyclate</a>
Maybell
coursework in korean coursework columbia courseware ku
Jolene
coursework based degrees java coursework coursework to order
Morris
coursework uitm coursework synonym coursework vs dissertation
Theo
coursework writing service coursework vs course work database coursework
Melanie
coursework vs dissertation coursework helper coursework writing help
Leave a comment
Your email address will not be published. Required fields are marked *